BRIGHT_NEIGHBOR,PERSIST_LOW
125-12
| 4176. | +/- | 2. |
| 4288. | +/- | 98. |
| 1.79 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -0.54 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.06 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.42 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 4420. | +/- | 5. |
| 4511. | +/- | 104. |
| 4.51 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.04 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW
125-12
| 4847. | +/- | 5. |
| 4906. | +/- | 91. |
| 2.77 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.42 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |

PERSIST_LOW
125-12
| 4270. | +/- | 1. |
| 4363. | +/- | 74. |
| 2.15 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.13 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.55 |
| -9999.00 |

PERSIST_LOW
125-12
| 3701. | +/- | 1. |
| 3779. | +/- | 60. |
| 1.15 | +/- | 0. |
| 0.96 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.57 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |
PERSIST_LOW
125-12
| 3873. | +/- | 1. |
| 3982. | +/- | 69. |
| 1.00 | +/- | 0. |
| 0.92 | +/- | 0. |
| 1.96 | +/- | 0. |
| 1.96 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.92 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.52 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 4418. | +/- | 5. |
| 4501. | +/- | 104. |
| 2.55 | +/- | 0. |
| 2.30 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.74 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.29 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW
125-12
| 4948. | +/- | 11. |
| 5001. | +/- | 123. |
| 2.71 | +/- | 0. |
| 2.39 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.77 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.74 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.49 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
125-12
| 8256. | +/- | 27. |
| 8093. | +/- | 266. |
| 3.86 | +/- | 0. |
| 3.81 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 53.11 | +/- | 0. |
| 1.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.90 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.22 |
| -9999.00 |
PERSIST_LOW
125-12
| 3909. | +/- | 1. |
| 3995. | +/- | 66. |
| 1.60 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.53 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 2.87 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.07 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW
STAR_WARN,COLORTE_WARN,SN_WARN
125-12
| 4038. | +/- | 5. |
| 4122. | +/- | 93. |
| 4.48 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.05 | +/- | 0. |
| 2.14 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.58 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW
125-12
| 5119. | +/- | 14. |
| 5147. | +/- | 127. |
| 3.50 | +/- | 0. |
| 3.22 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.62 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
125-12
| 4393. | +/- | 3. |
| 4496. | +/- | 91. |
| 2.12 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.65 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 5077. | +/- | 11. |
| 5119. | +/- | 101. |
| 3.13 | +/- | 0. |
| 3.01 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.92 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.03 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 4550. | +/- | 8. |
| 4629. | +/- | 109. |
| 4.52 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.78 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.29 |
| -9999.00 |
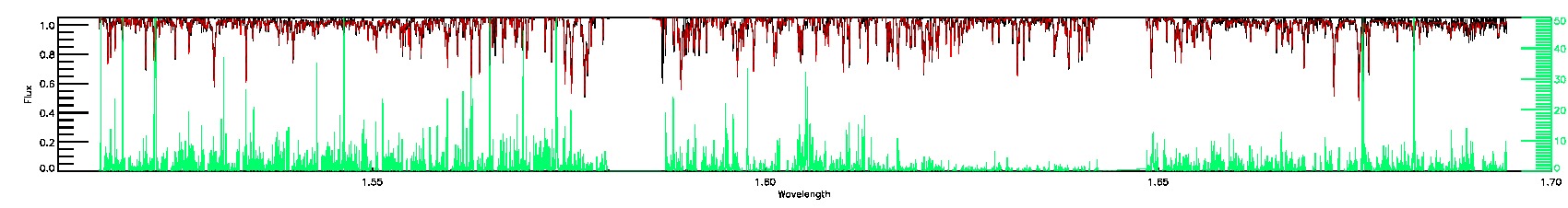
PERSIST_LOW
125-12
| 3690. | +/- | 1. |
| 3779. | +/- | 62. |
| 1.20 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.20 | +/- | 0. |
| 2.97 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 5154. | +/- | 28. |
| 5187. | +/- | 141. |
| 3.34 | +/- | 0. |
| 3.00 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.14 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.92 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.26 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
125-12
| 8478. | +/- | 32. |
| 8349. | +/- | 312. |
| 4.16 | +/- | 0. |
| 4.43 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 69.04 | +/- | 0. |
| 1.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.12 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.46 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 4726. | +/- | 8. |
| 4791. | +/- | 113. |
| 2.63 | +/- | 0. |
| 2.35 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.08 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.44 |
| -9999.00 |
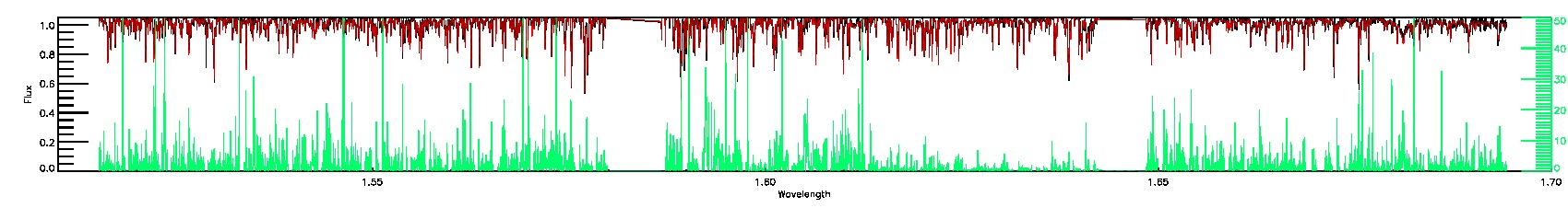
PERSIST_LOW
125-12
| 3895. | +/- | 1. |
| 3991. | +/- | 67. |
| 1.37 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.51 |
| -9999.00 |
PERSIST_LOW
125-12
| 4469. | +/- | 4. |
| 4558. | +/- | 96. |
| 4.36 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.04 | +/- | 0. |
| 7.68 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.34 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,COLORTE_WARN
125-12
| 4199. | +/- | 3. |
| 4296. | +/- | 81. |
| 4.34 | +/- | 0. |
| 4.63 | +/- | 0. |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.31 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
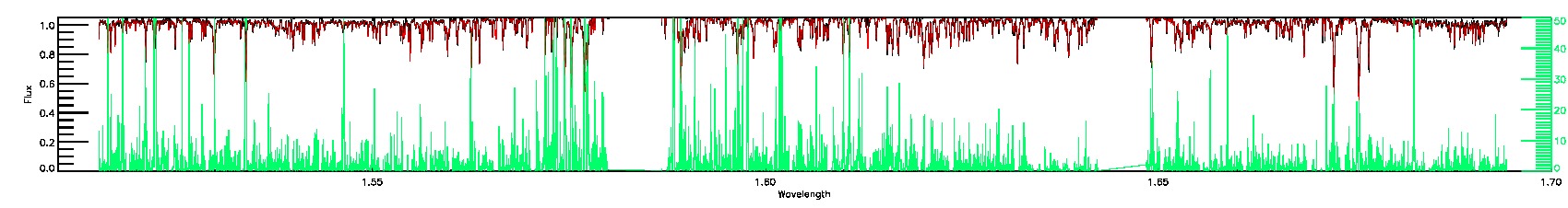
PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 4665. | +/- | 8. |
| 4733. | +/- | 111. |
| 2.84 | +/- | 0. |
| 2.63 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.91 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |

PERSIST_LOW
125-12
| 4214. | +/- | 1. |
| 4297. | +/- | 72. |
| 2.27 | +/- | 0. |
| 2.01 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.73 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.05 |
| -9999.00 |

125-12
| 4727. | +/- | 6. |
| 4771. | +/- | 93. |
| 3.52 | +/- | 0. |
| 3.31 | +/- | 0. |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.30 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.80 |
| -9999.00 |

125-12
| 4712. | +/- | 6. |
| 4774. | +/- | 96. |
| 3.04 | +/- | 0. |
| 2.84 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.91 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.29 |
| -9999.00 |
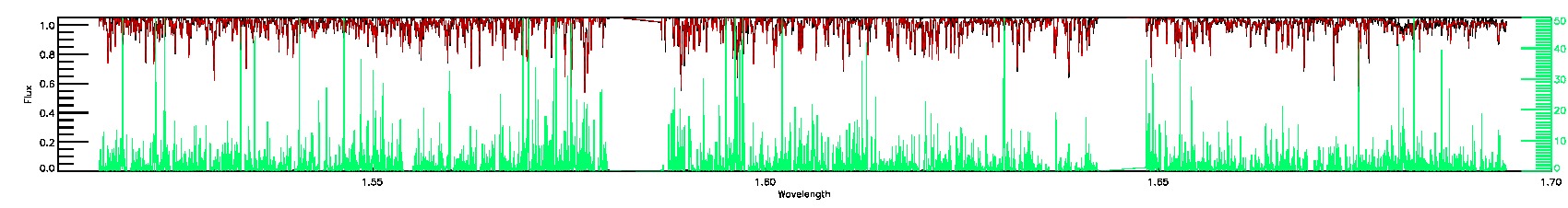
PERSIST_MED,PERSIST_LOW
125-12
| 4738. | +/- | 5. |
| 4812. | +/- | 88. |
| 2.70 | +/- | 0. |
| 2.39 | +/- | 0. |
| 1.55 | +/- | 0. |
| 1.55 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.56 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
PERSIST_LOW
125-12
| 4868. | +/- | 9. |
| 4933. | +/- | 111. |
| 2.68 | +/- | 0. |
| 2.37 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 3.83 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.46 |
| -9999.00 |

125-12
| 5046. | +/- | 16. |
| 5085. | +/- | 118. |
| 3.34 | +/- | 0. |
| 3.00 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.58 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.38 |
| -9999.00 |

125-12
| 3947. | +/- | 1. |
| 4058. | +/- | 71. |
| 1.33 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.83 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -0.53 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.03 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.92 |
| -9999.00 |
PERSIST_LOW
125-12
| 3628. | +/- | 1. |
| 3734. | +/- | 64. |
| 0.68 | +/- | 0. |
| 0.59 | +/- | 0. |
| 2.00 | +/- | 0. |
| 2.00 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.77 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.76 |
| -9999.00 |
125-12
| 3875. | +/- | 1. |
| 3988. | +/- | 70. |
| 1.12 | +/- | 0. |
| 1.04 | +/- | 0. |
| 2.04 | +/- | 0. |
| 2.04 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -0.57 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.13 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.84 |
| -9999.00 |
PERSIST_LOW
125-12
| 4250. | +/- | 2. |
| 4363. | +/- | 90. |
| 1.79 | +/- | 0. |
| 1.65 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -0.58 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.15 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.24 |
| -9999.00 |

125-12
| 4875. | +/- | 5. |
| 4929. | +/- | 90. |
| 3.11 | +/- | 0. |
| 2.95 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.34 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
125-12
| 8050. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 94.43 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.02 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.67 |
| -9999.00 |
PERSIST_LOW
STAR_BAD
STAR_WARN,COLORTE_WARN
125-12
| 7895. | +/- | 11. |
| -10000. | +/- | -NaN |
| 4.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.65 | +/- | 0. |
| 4.65 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 59.31 | +/- | 0. |
| 1.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -1.10 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW
125-12
| 4764. | +/- | 12. |
| 4830. | +/- | 115. |
| 2.94 | +/- | 0. |
| 2.57 | +/- | 0. |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.29 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -2.49 |
| -9999.00 |
125-12
| 3921. | +/- | 1. |
| 4003. | +/- | 65. |
| 1.75 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.09 | +/- | 0. |
| 2.71 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.01 |
| -9999.00 |
125-12
| 4618. | +/- | 4. |
| 4707. | +/- | 85. |
| 2.49 | +/- | 0. |
| 2.28 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.58 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.01 |
| -9999.00 |

125-12
| 4009. | +/- | 1. |
| 4083. | +/- | 65. |
| 1.89 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.44 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.36 |
| -9999.00 |
125-12
| 4148. | +/- | 1. |
| 4247. | +/- | 73. |
| 1.88 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.43 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 4834. | +/- | 12. |
| 4900. | +/- | 123. |
| 2.85 | +/- | 0. |
| 2.49 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 3.71 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.04 |
| -9999.00 |

125-12
| 4806. | +/- | 7. |
| 4852. | +/- | 99. |
| 3.42 | +/- | 0. |
| 3.24 | +/- | 0. |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.71 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.35 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
125-12
| 8298. | +/- | 30. |
| 8149. | +/- | 280. |
| 4.23 | +/- | 0. |
| 4.31 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 85.55 | +/- | 0. |
| 1.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.16 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.39 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
125-12
| 3682. | +/- | 1. |
| 3792. | +/- | 66. |
| -0.26 | +/- | 0. |
| -0.34 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.24 | +/- | 0. |
| 3.99 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.46 |
| -9999.00 |
125-12
| 3666. | +/- | 1. |
| 3772. | +/- | 65. |
| 0.94 | +/- | 0. |
| 0.84 | +/- | 0. |
| 1.89 | +/- | 0. |
| 1.89 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.27 | +/- | 0. |
| 3.78 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.22 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.04 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 8608. | +/- | 43. |
| 8475. | +/- | 342. |
| 4.21 | +/- | 0. |
| 4.45 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.34 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.79 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.45 |
| -9999.00 |
125-12
| 4154. | +/- | 1. |
| 4233. | +/- | 69. |
| 2.17 | +/- | 0. |
| 1.91 | +/- | 0. |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.62 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.89 |
| -9999.00 |
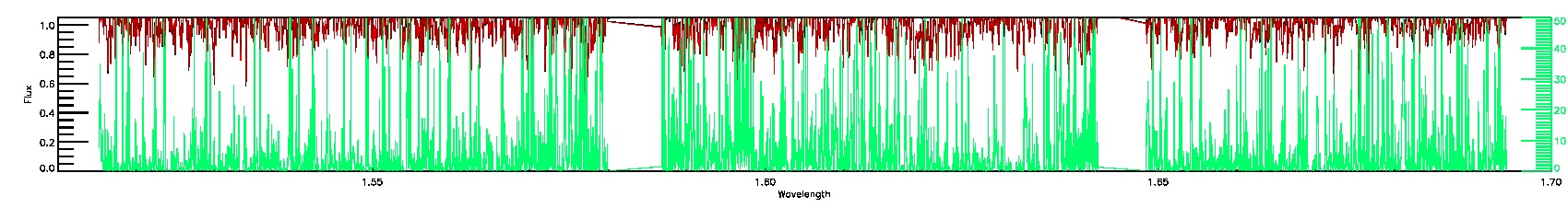
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 11281. | +/- | 103. |
| -10000. | +/- | -NaN |
| 4.46 | +/- | 0. |
| 4.44 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.39 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.96 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.97 |
| -9999.00 |
125-12
| 3969. | +/- | 1. |
| 4052. | +/- | 66. |
| 1.74 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.76 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.40 |
| -9999.00 |
125-12
| 4443. | +/- | 2. |
| 4539. | +/- | 79. |
| 2.42 | +/- | 0. |
| 2.21 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |

125-12
| 4666. | +/- | 7. |
| 4738. | +/- | 105. |
| 2.84 | +/- | 0. |
| 2.64 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.08 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.04 |
| -9999.00 |

PERSIST_LOW
125-12
| 4577. | +/- | 3. |
| 4640. | +/- | 85. |
| 3.17 | +/- | 0. |
| 2.94 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.37 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.43 |
| -9999.00 |

PERSIST_LOW
125-12
| 3949. | +/- | 1. |
| 4051. | +/- | 69. |
| 1.39 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.57 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.59 |
| -9999.00 |
125-12
| 4613. | +/- | 4. |
| 4695. | +/- | 83. |
| 2.75 | +/- | 0. |
| 2.56 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.25 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.32 |
| -9999.00 |
125-12
| 5048. | +/- | 7. |
| 5079. | +/- | 94. |
| 3.10 | +/- | 0. |
| 2.72 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.25 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.19 |
| -9999.00 |

125-12
| 3911. | +/- | 1. |
| 4031. | +/- | 72. |
| 1.14 | +/- | 0. |
| 1.08 | +/- | 0. |
| 2.10 | +/- | 0. |
| 2.10 | +/- | -NaN |
| -0.72 | +/- | 0. |
| -0.72 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.88 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4911. | +/- | 6. |
| 4963. | +/- | 99. |
| 2.78 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.44 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.11 |
| -9999.00 |
PERSIST_MED
125-12
| 4705. | +/- | 4. |
| 4772. | +/- | 84. |
| 2.75 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.05 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.04 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4092. | +/- | 2. |
| 4205. | +/- | 91. |
| 1.56 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -0.57 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.14 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -1.44 |
| -9999.00 |

125-12
| 3708. | +/- | 1. |
| 3821. | +/- | 67. |
| 0.71 | +/- | 0. |
| 0.63 | +/- | 0. |
| 2.06 | +/- | 0. |
| 2.06 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -0.58 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.82 |
| -9999.00 |
PERSIST_MED
125-12
| 4923. | +/- | 6. |
| 4979. | +/- | 93. |
| 2.76 | +/- | 0. |
| 2.43 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.62 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.72 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION
125-12
| 8347. | +/- | 27. |
| 8196. | +/- | 282. |
| 4.28 | +/- | 0. |
| 4.34 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 20.74 | +/- | 0. |
| 1.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.20 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.04 |
| -9999.00 |
125-12
| 3858. | +/- | 1. |
| 3954. | +/- | 66. |
| 1.33 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.29 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.31 |
| -9999.00 |
PERSIST_LOW
125-12
| 5040. | +/- | 9. |
| 5057. | +/- | 114. |
| 4.49 | +/- | 0. |
| 4.46 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 1.77 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.19 |
| -9999.00 |

PERSIST_LOW
125-12
| 3938. | +/- | 1. |
| 4046. | +/- | 70. |
| 1.26 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.88 | +/- | 0. |
| 1.88 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.87 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.58 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
125-12
| 8281. | +/- | 24. |
| 8161. | +/- | 302. |
| 4.40 | +/- | 0. |
| 4.77 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 33.35 | +/- | 0. |
| 1.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.30 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.06 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD
125-12
| 7999. | +/- | 12. |
| -10000. | +/- | -NaN |
| 4.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| -1.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.83 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.31 |
| -9999.00 |
STAR_WARN,SN_WARN
125-12
| 4744. | +/- | 10. |
| 4815. | +/- | 117. |
| 2.49 | +/- | 0. |
| 2.28 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.42 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.51 |
| -9999.00 |
125-12
| 4659. | +/- | 4. |
| 4735. | +/- | 84. |
| 2.87 | +/- | 0. |
| 2.68 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.37 |
| -9999.00 |
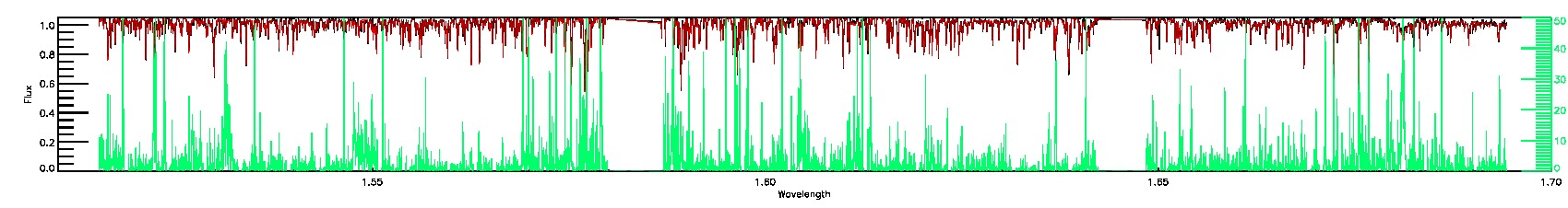
BRIGHT_NEIGHBOR,PERSIST_LOW
125-12
| 4398. | +/- | 4. |
| 4517. | +/- | 98. |
| 2.01 | +/- | 0. |
| 1.87 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.58 | +/- | -NaN |
| -0.70 | +/- | 0. |
| -0.70 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.34 |
| -9999.00 |

PERSIST_LOW
125-12
| 4819. | +/- | 5. |
| 4878. | +/- | 88. |
| 2.99 | +/- | 0. |
| 2.82 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
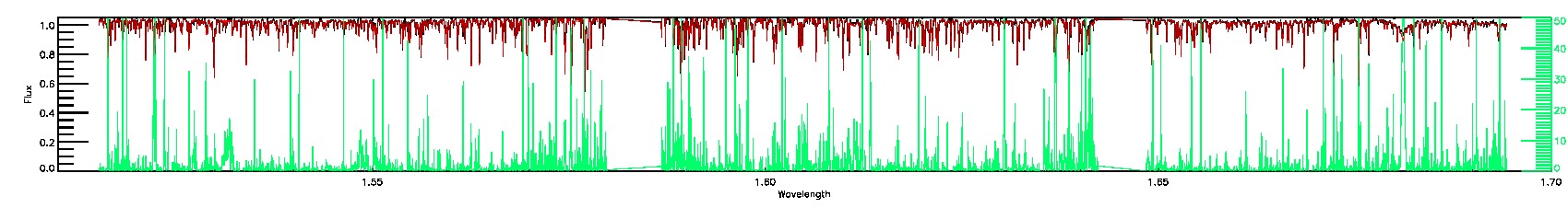
125-12
| 4896. | +/- | 7. |
| 4955. | +/- | 92. |
| 2.73 | +/- | 0. |
| 2.41 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.68 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.38 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.69 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.48 |
| -9999.00 |
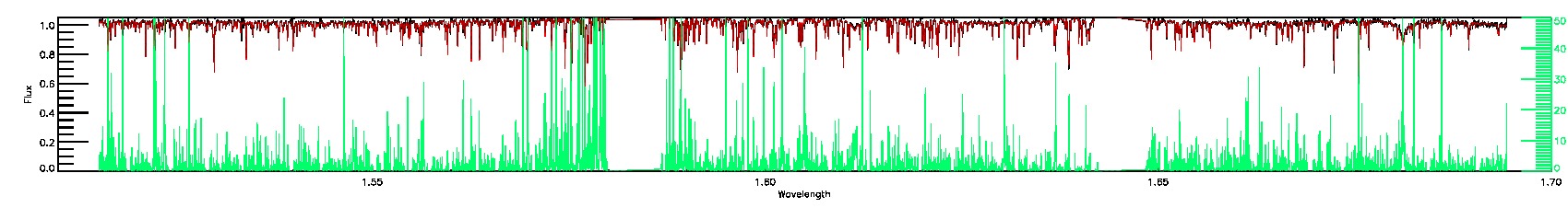
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 7880. | +/- | 22. |
| 7703. | +/- | 229. |
| 4.15 | +/- | 0. |
| 3.97 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 34.47 | +/- | 0. |
| 1.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.85 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.24 |
| -9999.00 |
125-12
| 4478. | +/- | 5. |
| 4584. | +/- | 100. |
| 2.28 | +/- | 0. |
| 2.10 | +/- | 0. |
| 1.55 | +/- | 0. |
| 1.55 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.79 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.65 |
| -9999.00 |

PERSIST_MED,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
125-12
| 5841. | +/- | 36. |
| -10000. | +/- | -NaN |
| 3.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.20 | +/- | 1. |
| 1.20 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.37 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.67 |
| -9999.00 |
125-12
| 4080. | +/- | 1. |
| 4174. | +/- | 71. |
| 1.84 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.17 | +/- | 0. |
| 3.19 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.34 |
| -9999.00 |
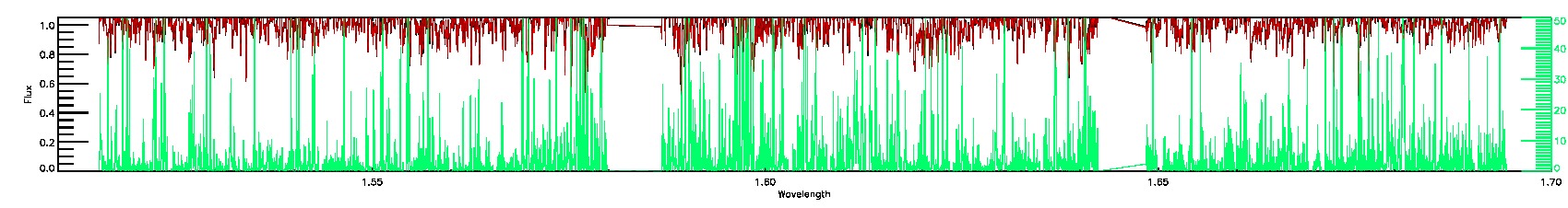
125-12
| 4236. | +/- | 3. |
| 4325. | +/- | 87. |
| 4.36 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.66 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.05 |
| -9999.00 |

PERSIST_LOW
125-12
| 4037. | +/- | 1. |
| 4132. | +/- | 70. |
| 1.71 | +/- | 0. |
| 1.52 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.25 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.48 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 5016. | +/- | 11. |
| 5044. | +/- | 108. |
| 3.37 | +/- | 0. |
| 3.05 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.95 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
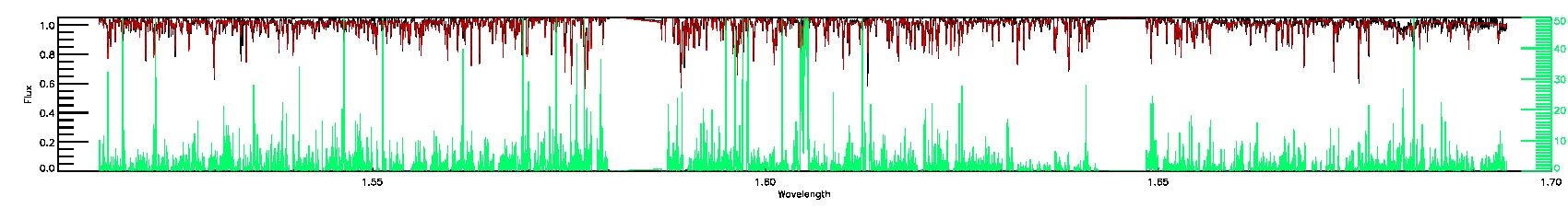
STAR_WARN,COLORTE_WARN
125-12
| 4445. | +/- | 3. |
| 4538. | +/- | 84. |
| 4.19 | +/- | 0. |
| 4.39 | +/- | 0. |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.40 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.11 |
| -9999.00 |

PERSIST_LOW
125-12
| 4038. | +/- | 1. |
| 4122. | +/- | 68. |
| 1.87 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.77 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |

PERSIST_LOW
125-12
| 3816. | +/- | 1. |
| 3907. | +/- | 65. |
| 1.30 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.72 | +/- | 0. |
| 1.72 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.04 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
125-12
| 4538. | +/- | 3. |
| 4623. | +/- | 81. |
| 2.87 | +/- | 0. |
| 2.66 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.10 | +/- | 0. |
| 3.00 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.12 |
| -9999.00 |
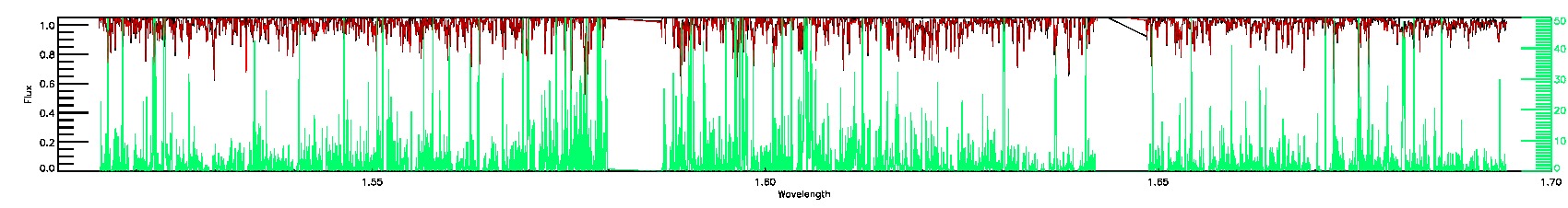
BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
125-12
| 8394. | +/- | 34. |
| 8266. | +/- | 312. |
| 4.18 | +/- | 0. |
| 4.46 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.73 | +/- | 0. |
| 1.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.88 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.59 |
| -9999.00 |
125-12
| 4934. | +/- | 8. |
| 4984. | +/- | 102. |
| 2.80 | +/- | 0. |
| 2.46 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.50 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.29 |
| -9999.00 |

SUSPECT_RV_COMBINATION
125-12
| 8406. | +/- | 29. |
| 8295. | +/- | 321. |
| 4.42 | +/- | 0. |
| 4.88 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 26.65 | +/- | 0. |
| 1.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.35 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -2.13 |
| -9999.00 |
PERSIST_LOW
125-12
| 4919. | +/- | 7. |
| 4958. | +/- | 112. |
| 3.27 | +/- | 0. |
| 2.91 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.92 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.16 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 5056. | +/- | 7. |
| 5086. | +/- | 94. |
| 3.65 | +/- | 0. |
| 3.58 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.24 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
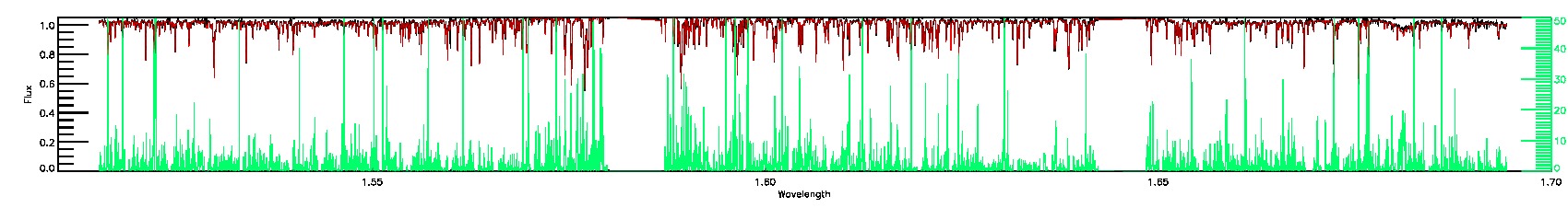
125-12
| 4768. | +/- | 6. |
| 4834. | +/- | 92. |
| 2.92 | +/- | 0. |
| 2.75 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.33 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.98 |
| -9999.00 |
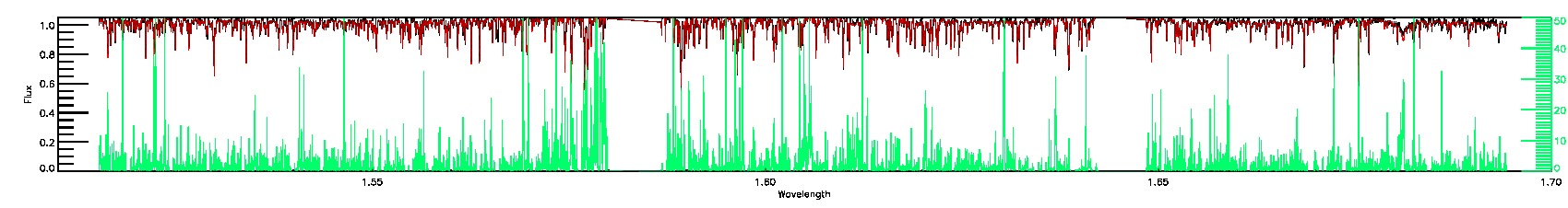
125-12
| 4855. | +/- | 6. |
| 4906. | +/- | 88. |
| 3.29 | +/- | 0. |
| 3.13 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.15 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
STAR_WARN,SN_WARN
125-12
| 4584. | +/- | 7. |
| 4671. | +/- | 111. |
| 2.63 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.34 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.08 |
| -9999.00 |

SUSPECT_RV_COMBINATION
125-12
| 8541. | +/- | 31. |
| 8432. | +/- | 336. |
| 4.24 | +/- | 0. |
| 4.72 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.00 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.32 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -2.30 |
| -9999.00 |
125-12
| 4147. | +/- | 1. |
| 4260. | +/- | 76. |
| 1.61 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -0.57 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.13 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.93 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4826. | +/- | 6. |
| 4888. | +/- | 92. |
| 2.68 | +/- | 0. |
| 2.37 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.48 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.07 |
| -9999.00 |
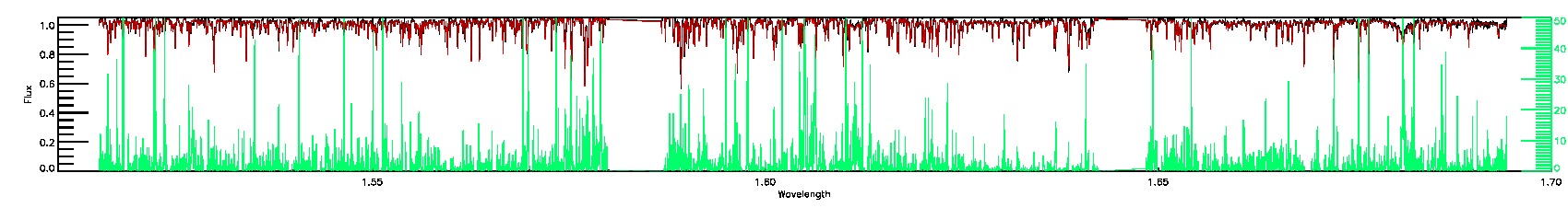
125-12
| 4357. | +/- | 2. |
| 4450. | +/- | 76. |
| 4.44 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.82 | +/- | 0. |
| 0.82 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 5.02 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.31 |
| -9999.00 |

SUSPECT_RV_COMBINATION
125-12
| 8529. | +/- | 30. |
| 8427. | +/- | 341. |
| 4.29 | +/- | 0. |
| 4.82 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 24.81 | +/- | 0. |
| 1.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.34 |
| -9999.00 |
125-12
| 4259. | +/- | 1. |
| 4355. | +/- | 75. |
| 1.97 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.47 |
| -9999.00 |
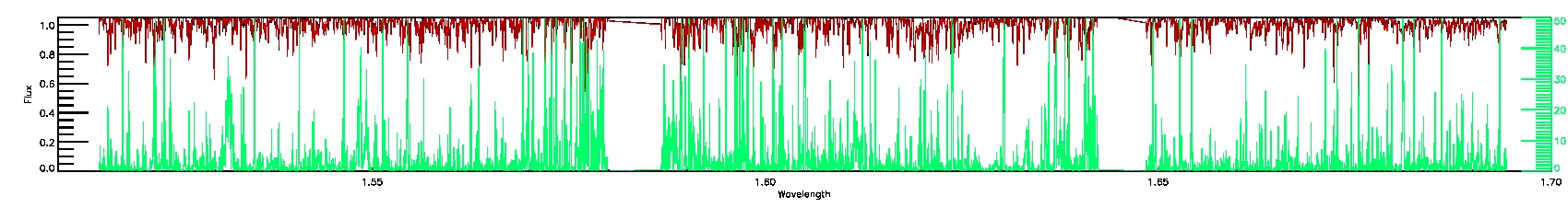
STAR_WARN,COLORTE_WARN
125-12
| 4019. | +/- | 3. |
| 4118. | +/- | 87. |
| 4.32 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.48 | +/- | 0. |
| 0.48 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.53 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.32 |
| -9999.00 |
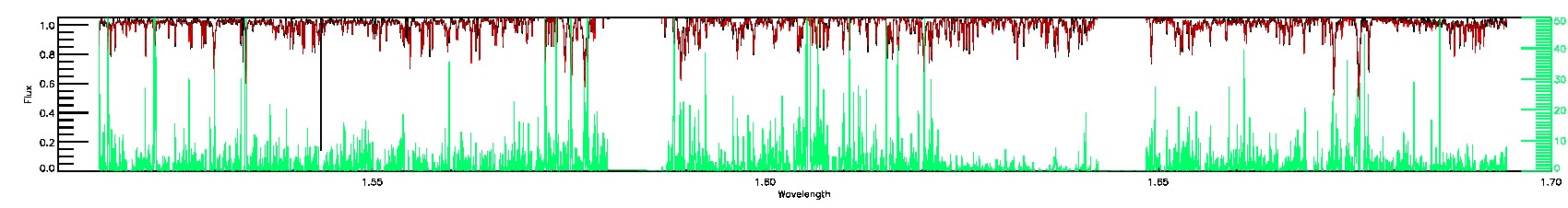
125-12
| 4351. | +/- | 3. |
| 4461. | +/- | 86. |
| 4.34 | +/- | 0. |
| 4.72 | +/- | 0. |
| 0.73 | +/- | 0. |
| 0.73 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.39 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.45 |
| -9999.00 |

125-12
| 4808. | +/- | 5. |
| 4859. | +/- | 86. |
| 3.15 | +/- | 0. |
| 2.96 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.88 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.18 |
| -9999.00 |

PERSIST_LOW
125-12
| 3672. | +/- | 1. |
| 3786. | +/- | 66. |
| 0.63 | +/- | 0. |
| 0.55 | +/- | 0. |
| 2.14 | +/- | 0. |
| 2.14 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -0.59 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.18 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.25 |
| -9999.00 |
125-12
| 4635. | +/- | 3. |
| 4694. | +/- | 79. |
| 3.10 | +/- | 0. |
| 2.87 | +/- | 0. |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.44 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.96 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
125-12
| 4920. | +/- | 8. |
| 4979. | +/- | 108. |
| 2.91 | +/- | 0. |
| 2.55 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.87 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.32 |
| -9999.00 |
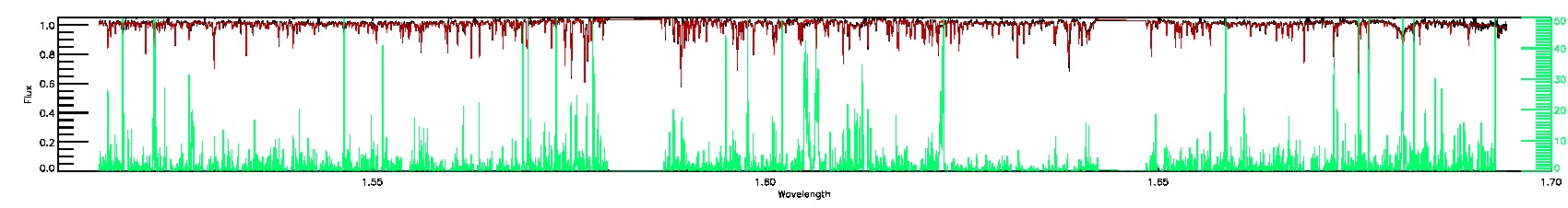
125-12
| 3834. | +/- | 1. |
| 3927. | +/- | 65. |
| 1.29 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.16 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
125-12
| 4729. | +/- | 10. |
| 4811. | +/- | 110. |
| 2.65 | +/- | 0. |
| 2.49 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.89 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.66 |
| -9999.00 |
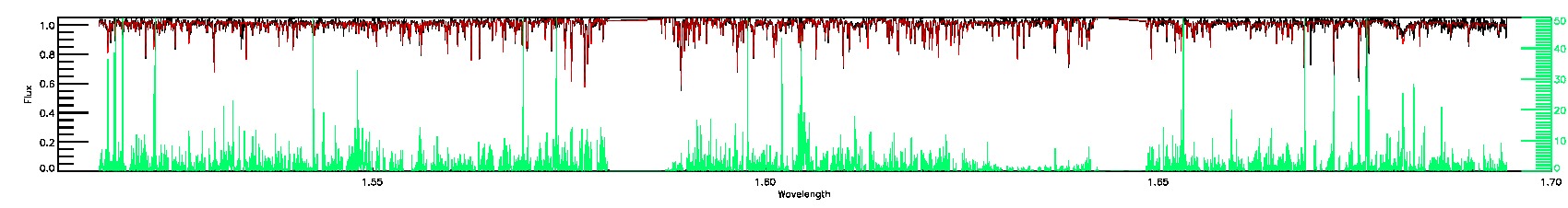
125-12
| 3938. | +/- | 1. |
| 4046. | +/- | 70. |
| 1.20 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.97 | +/- | 0. |
| 1.97 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.86 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4483. | +/- | 3. |
| 4575. | +/- | 79. |
| 2.53 | +/- | 0. |
| 2.32 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.13 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.53 |
| -9999.00 |
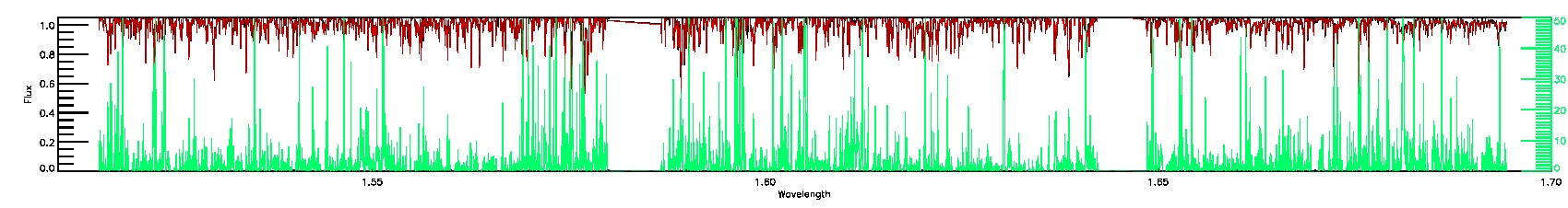
125-12
| 3951. | +/- | 1. |
| 4044. | +/- | 68. |
| 1.57 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.17 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.29 |
| -9999.00 |
125-12
| 4295. | +/- | 2. |
| 4379. | +/- | 73. |
| 2.21 | +/- | 0. |
| 1.96 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.82 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.27 |
| -9999.00 |

125-12
| 3946. | +/- | 1. |
| 4045. | +/- | 69. |
| 1.46 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.51 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.25 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.43 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.84 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4921. | +/- | 9. |
| 4948. | +/- | 104. |
| 3.62 | +/- | 0. |
| 3.47 | +/- | 0. |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.47 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.08 |
| -9999.00 |

125-12
| 5072. | +/- | 6. |
| 5093. | +/- | 93. |
| 3.27 | +/- | 0. |
| 2.92 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.89 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.32 |
| -9999.00 |
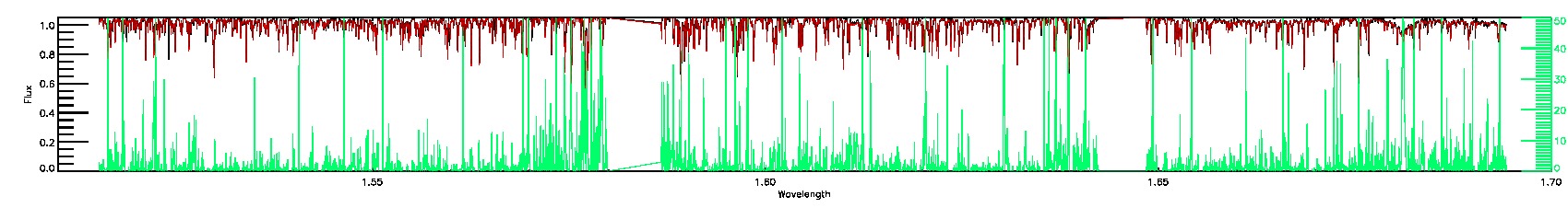
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
125-12
| 7999. | +/- | 13. |
| -10000. | +/- | -NaN |
| 4.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.95 | +/- | 0. |
| 0.95 | +/- | -NaN |
| -2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.58 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.24 |
| -9999.00 |
125-12
| 4026. | +/- | 1. |
| 4109. | +/- | 67. |
| 1.91 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.74 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4314. | +/- | 2. |
| 4421. | +/- | 78. |
| 2.05 | +/- | 0. |
| 1.87 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 3.81 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.59 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
125-12
| 4808. | +/- | 9. |
| 4866. | +/- | 110. |
| 2.65 | +/- | 0. |
| 2.36 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.14 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| 3.19 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.14 |
| -9999.00 |

125-12
| 3796. | +/- | 1. |
| 3890. | +/- | 65. |
| 1.37 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.21 | +/- | 0. |
| 3.19 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.24 |
| -9999.00 |
125-12
| 4889. | +/- | 7. |
| 4954. | +/- | 94. |
| 2.66 | +/- | 0. |
| 2.36 | +/- | 0. |
| 1.78 | +/- | 0. |
| 1.78 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.10 | +/- | 0. |
| 3.98 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.08 |
| -9999.00 |
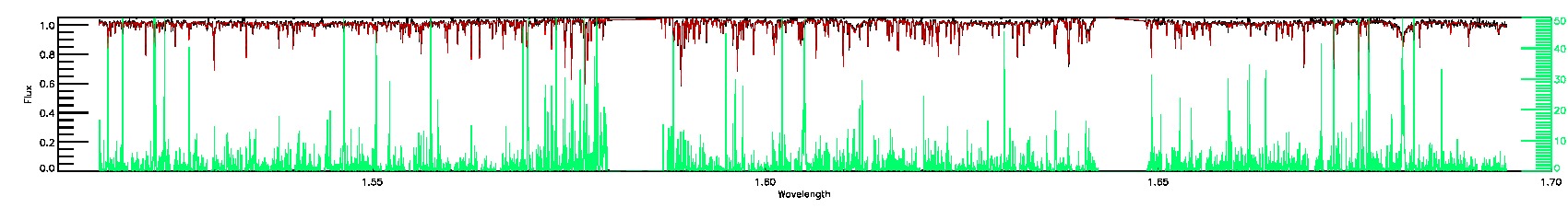
PERSIST_LOW
125-12
| 3877. | +/- | 1. |
| 3993. | +/- | 71. |
| 1.02 | +/- | 0. |
| 0.96 | +/- | 0. |
| 1.95 | +/- | 0. |
| 1.95 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -0.66 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.11 |
| -9999.00 |
PERSIST_LOW
125-12
| 3979. | +/- | 1. |
| 4086. | +/- | 71. |
| 1.40 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.83 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.78 |
| -9999.00 |
125-12
| 3920. | +/- | 1. |
| 4041. | +/- | 72. |
| 1.13 | +/- | 0. |
| 1.08 | +/- | 0. |
| 2.15 | +/- | 0. |
| 2.15 | +/- | -NaN |
| -0.77 | +/- | 0. |
| -0.77 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.21 |
| -9999.00 |
125-12
| 4514. | +/- | 3. |
| 4606. | +/- | 91. |
| 2.77 | +/- | 0. |
| 2.57 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.15 | +/- | 0. |
| 3.18 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.34 |
| -9999.00 |
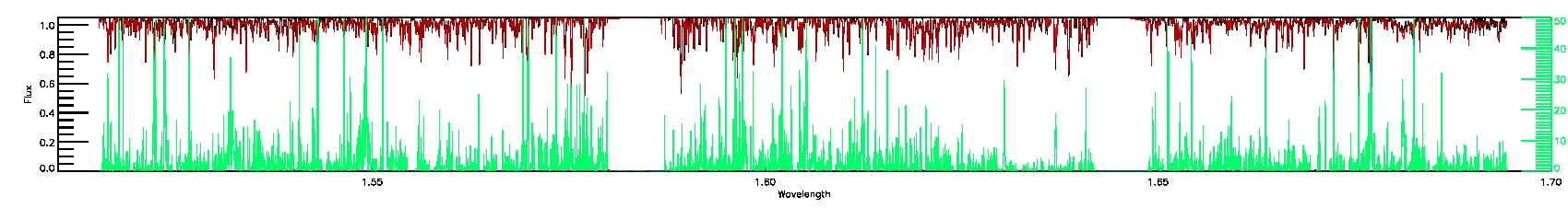
SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 8984. | +/- | 59. |
| 8865. | +/- | 372. |
| 3.97 | +/- | 0. |
| 4.34 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 69.10 | +/- | 0. |
| 1.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.94 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.08 |
| -9999.00 |
PERSIST_LOW
125-12
| 4052. | +/- | 1. |
| 4139. | +/- | 69. |
| 1.82 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.93 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
125-12
| 7996. | +/- | 12. |
| -10000. | +/- | -NaN |
| 4.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.74 | +/- | 1. |
| 0.74 | +/- | -NaN |
| -2.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.77 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.16 |
| -9999.00 |
125-12
| 4212. | +/- | 1. |
| 4321. | +/- | 76. |
| 1.85 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.91 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.92 |
| -9999.00 |
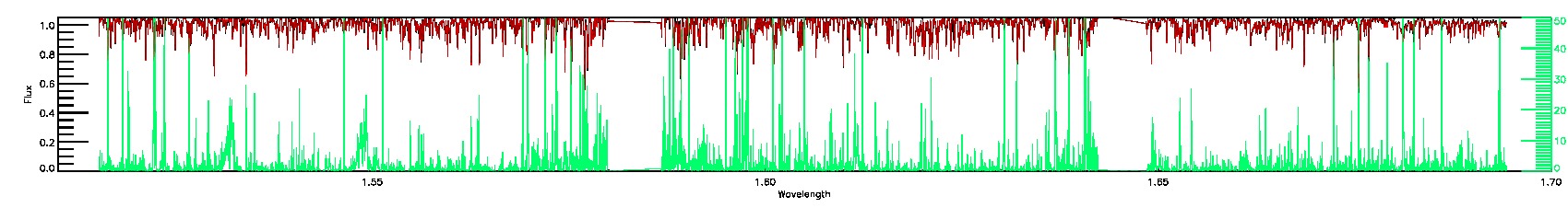
125-12
| 3909. | +/- | 1. |
| 4016. | +/- | 69. |
| 1.24 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.90 | +/- | 0. |
| 1.90 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.81 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.31 |
| -9999.00 |
125-12
| 3593. | +/- | 1. |
| 3718. | +/- | 66. |
| 0.40 | +/- | 0. |
| 0.37 | +/- | 0. |
| 2.56 | +/- | 0. |
| 2.56 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -0.86 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.22 | +/- | 0. |
| 4.89 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.53 |
| -9999.00 |
125-12
| 4990. | +/- | 9. |
| 5029. | +/- | 103. |
| 3.42 | +/- | 0. |
| 3.28 | +/- | 0. |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.30 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |

125-12
| 4117. | +/- | 1. |
| 4228. | +/- | 75. |
| 1.62 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -0.52 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.01 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.38 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4236. | +/- | 2. |
| 4341. | +/- | 87. |
| 1.93 | +/- | 0. |
| 1.75 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.73 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.26 |
| -9999.00 |

PERSIST_LOW
125-12
| 3990. | +/- | 1. |
| 4085. | +/- | 69. |
| 1.56 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.60 | +/- | 0. |
| 1.60 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.26 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.20 |
| -9999.00 |

PERSIST_LOW
125-12
| 4796. | +/- | 6. |
| 4859. | +/- | 100. |
| 2.69 | +/- | 0. |
| 2.38 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.62 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.34 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.63 |
| -9999.00 |

125-12
| 4616. | +/- | 3. |
| 4690. | +/- | 81. |
| 2.69 | +/- | 0. |
| 2.38 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.91 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.26 |
| -9999.00 |

SUSPECT_RV_COMBINATION
125-12
| 8239. | +/- | 23. |
| 8117. | +/- | 296. |
| 4.44 | +/- | 0. |
| 4.79 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.05 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.03 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -2.02 |
| -9999.00 |
125-12
| 5035. | +/- | 11. |
| 5078. | +/- | 107. |
| 3.12 | +/- | 0. |
| 2.74 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.38 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.70 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.26 |
| -9999.00 |
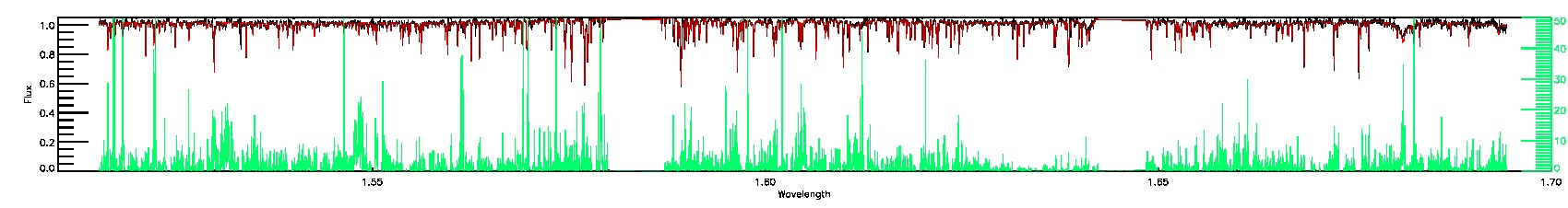
BRIGHT_NEIGHBOR
125-12
| 4779. | +/- | 7. |
| 4836. | +/- | 102. |
| 2.75 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.99 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.01 |
| -9999.00 |

PERSIST_LOW
STAR_WARN,COLORTE_WARN
125-12
| 8653. | +/- | 44. |
| 8536. | +/- | 363. |
| 4.08 | +/- | 0. |
| 4.48 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.72 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.28 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -1.86 |
| -9999.00 |
125-12
| 4738. | +/- | 7. |
| 4805. | +/- | 102. |
| 2.74 | +/- | 0. |
| 2.41 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.20 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.57 |
| -9999.00 |
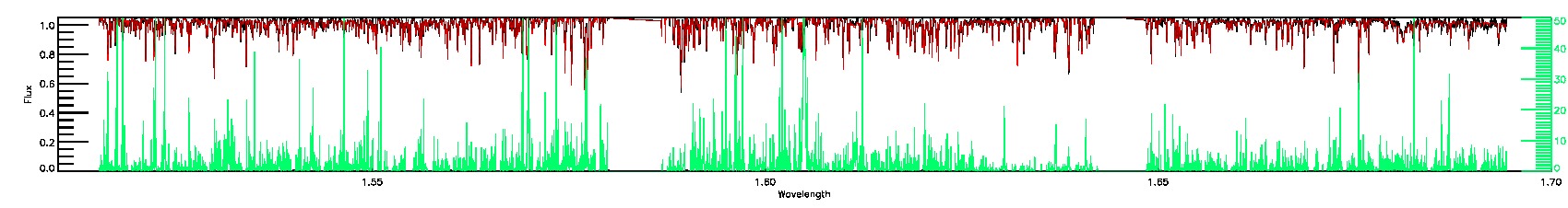
125-12
| 4347. | +/- | 2. |
| 4428. | +/- | 74. |
| 2.55 | +/- | 0. |
| 2.30 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 2.67 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.24 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4960. | +/- | 6. |
| 5001. | +/- | 92. |
| 2.91 | +/- | 0. |
| 2.54 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.17 |
| -9999.00 |

125-12
| 4442. | +/- | 3. |
| 4529. | +/- | 85. |
| 2.57 | +/- | 0. |
| 2.34 | +/- | 0. |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.90 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.30 |
| -9999.00 |
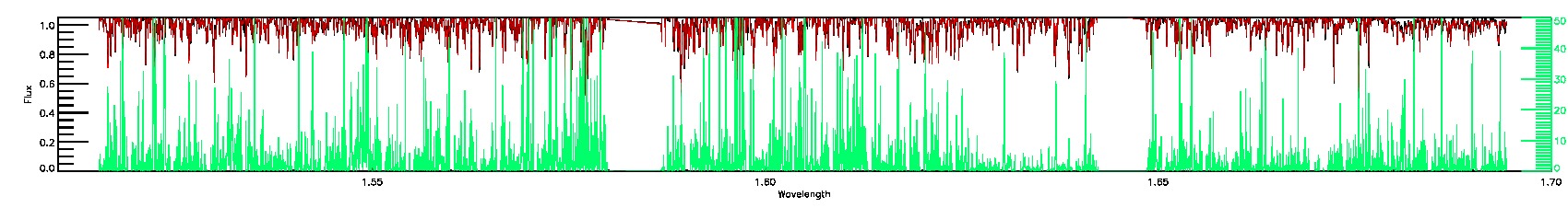
125-12
| 4066. | +/- | 1. |
| 4166. | +/- | 71. |
| 1.65 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.49 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.66 |
| -9999.00 |

PERSIST_LOW
STAR_WARN,COLORTE_WARN
125-12
| 3688. | +/- | 1. |
| 3805. | +/- | 67. |
| 0.61 | +/- | 0. |
| 0.55 | +/- | 0. |
| 2.33 | +/- | 0. |
| 2.33 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -0.67 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.11 | +/- | 0. |
| 4.37 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.72 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4907. | +/- | 9. |
| 4964. | +/- | 107. |
| 2.77 | +/- | 0. |
| 2.43 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.65 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.68 |
| -9999.00 |
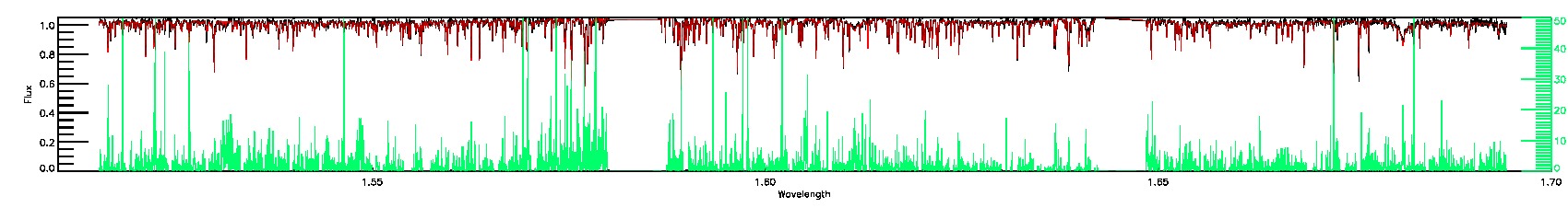
PERSIST_LOW
125-12
| 3832. | +/- | 1. |
| 3947. | +/- | 69. |
| 0.97 | +/- | 0. |
| 0.90 | +/- | 0. |
| 1.96 | +/- | 0. |
| 1.96 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -0.61 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.10 | +/- | 0. |
| 4.23 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.97 |
| -9999.00 |
125-12
| 4526. | +/- | 4. |
| 4611. | +/- | 87. |
| 4.44 | +/- | 0. |
| 4.57 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.09 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |

125-12
| 4758. | +/- | 4. |
| 4815. | +/- | 85. |
| 2.85 | +/- | 0. |
| 2.49 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.89 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.05 |
| -9999.00 |

125-12
| 4898. | +/- | 7. |
| 4952. | +/- | 100. |
| 3.01 | +/- | 0. |
| 2.63 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.49 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.60 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4681. | +/- | 5. |
| 4749. | +/- | 83. |
| 2.93 | +/- | 0. |
| 2.73 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.98 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |

125-12
| 4897. | +/- | 6. |
| 4959. | +/- | 93. |
| 2.68 | +/- | 0. |
| 2.38 | +/- | 0. |
| 1.72 | +/- | 0. |
| 1.72 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.84 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.14 |
| -9999.00 |

125-12
| 4421. | +/- | 2. |
| 4502. | +/- | 75. |
| 2.49 | +/- | 0. |
| 2.24 | +/- | 0. |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.67 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.17 |
| -9999.00 |
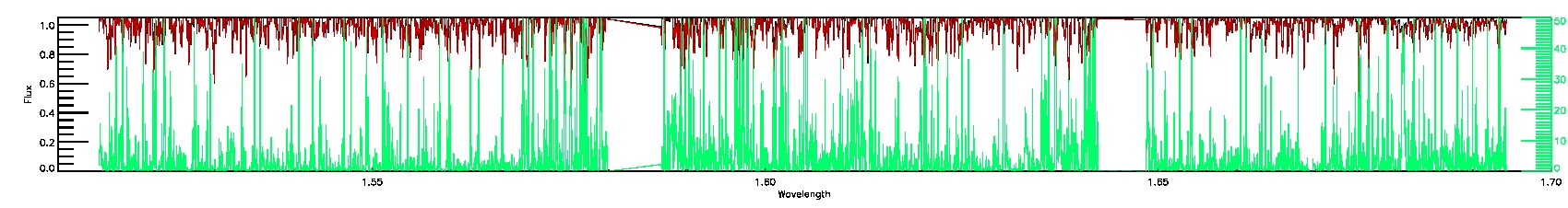
125-12
| 4131. | +/- | 1. |
| 4228. | +/- | 74. |
| 1.83 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.58 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.20 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.33 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.41 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
125-12
| 4869. | +/- | 9. |
| 4921. | +/- | 107. |
| 4.55 | +/- | 0. |
| 4.70 | +/- | 0. |
| 0.75 | +/- | 0. |
| 0.75 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.02 |
| -9999.00 |

125-12
| 3862. | +/- | 1. |
| 3958. | +/- | 66. |
| 1.31 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.40 |
| -9999.00 |
125-12
| 4004. | +/- | 1. |
| 4112. | +/- | 72. |
| 0.79 | +/- | 0. |
| 0.70 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.54 | +/- | 0. |
| 0.54 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.07 | +/- | 0. |
| 3.87 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.30 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_BAD
125-12
| 7224. | +/- | 18. |
| -10000. | +/- | -NaN |
| 3.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.94 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.33 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
125-12
| 3768. | +/- | 1. |
| 3899. | +/- | 71. |
| 0.56 | +/- | 0. |
| 0.54 | +/- | 0. |
| 2.15 | +/- | 0. |
| 2.15 | +/- | -NaN |
| -0.99 | +/- | 0. |
| -0.99 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 5.27 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.87 |
| -9999.00 |
125-12
| 4765. | +/- | 4. |
| 4827. | +/- | 86. |
| 2.76 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.14 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.27 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4956. | +/- | 10. |
| 5005. | +/- | 110. |
| 2.77 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |

125-12
| 4730. | +/- | 7. |
| 4822. | +/- | 98. |
| 2.51 | +/- | 0. |
| 2.28 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -0.70 | +/- | 0. |
| -0.70 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.12 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.95 |
| -9999.00 |
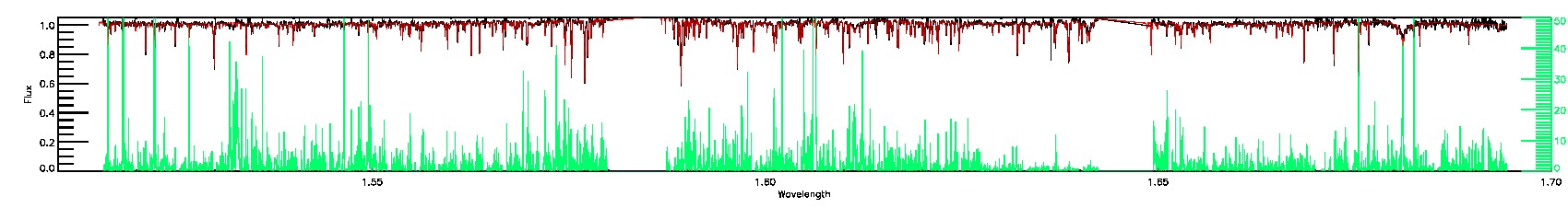
BRIGHT_NEIGHBOR
125-12
| 4928. | +/- | 10. |
| 4972. | +/- | 107. |
| 3.65 | +/- | 0. |
| 3.60 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.11 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.11 | +/- | 0. |
| 3.16 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.19 |
| -9999.00 |
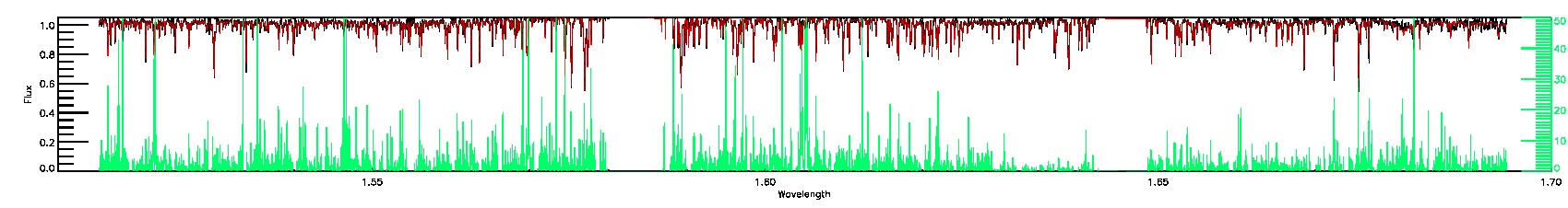
125-12
| 3547. | +/- | 1. |
| 3660. | +/- | 63. |
| 0.35 | +/- | 0. |
| 0.28 | +/- | 0. |
| 2.15 | +/- | 0. |
| 2.15 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -0.57 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.13 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.91 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
125-12
| 4446. | +/- | 4. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.85 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -0.22 |
| -9999.00 |

125-12
| 3904. | +/- | 1. |
| 4011. | +/- | 69. |
| 1.13 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.79 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.83 |
| -9999.00 |
PERSIST_LOW
125-12
| 3763. | +/- | 1. |
| 3859. | +/- | 65. |
| 1.07 | +/- | 0. |
| 0.94 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.29 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.49 |
| -9999.00 |
125-12
| 4198. | +/- | 1. |
| 4289. | +/- | 72. |
| 1.95 | +/- | 0. |
| 1.72 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.05 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.58 |
| -9999.00 |

PERSIST_LOW
125-12
| 4183. | +/- | 3. |
| 4296. | +/- | 96. |
| 1.80 | +/- | 0. |
| 1.65 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -0.57 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.12 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.07 |
| -9999.00 |

125-12
| 3897. | +/- | 1. |
| 3992. | +/- | 67. |
| 1.40 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.24 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
125-12
| 3574. | +/- | 1. |
| 3686. | +/- | 64. |
| 0.64 | +/- | 0. |
| 0.56 | +/- | 0. |
| 2.27 | +/- | 0. |
| 2.27 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -0.54 | +/- | 0. |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.26 | +/- | 0. |
| 4.05 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.62 |
| -9999.00 |
125-12
| 3748. | +/- | 1. |
| 3861. | +/- | 67. |
| 0.79 | +/- | 0. |
| 0.71 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.93 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -0.56 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.10 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.56 |
| -9999.00 |
125-12
| 4064. | +/- | 1. |
| 4145. | +/- | 68. |
| 1.68 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.67 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.23 |
| -9999.00 |

PERSIST_LOW
125-12
| 3891. | +/- | 1. |
| 3972. | +/- | 64. |
| 1.70 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.55 | +/- | 0. |
| 1.55 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.65 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.14 |
| -9999.00 |
125-12
| 3866. | +/- | 1. |
| 3987. | +/- | 71. |
| 0.99 | +/- | 0. |
| 0.94 | +/- | 0. |
| 2.01 | +/- | 0. |
| 2.01 | +/- | -NaN |
| -0.76 | +/- | 0. |
| -0.76 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.01 |
| -9999.00 |
125-12
| 4889. | +/- | 6. |
| 4957. | +/- | 94. |
| 2.64 | +/- | 0. |
| 2.35 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -0.58 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 4.15 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.89 |
| -9999.00 |

PERSIST_LOW
125-12
| 3645. | +/- | 1. |
| 3756. | +/- | 65. |
| 0.60 | +/- | 0. |
| 0.52 | +/- | 0. |
| 2.13 | +/- | 0. |
| 2.13 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -0.53 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.02 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.03 |
| -9999.00 |
125-12
| 3920. | +/- | 1. |
| 4028. | +/- | 70. |
| 1.19 | +/- | 0. |
| 1.10 | +/- | 0. |
| 1.86 | +/- | 0. |
| 1.86 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.88 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.31 |
| -9999.00 |
PERSIST_LOW
125-12
| 5006. | +/- | 9. |
| 5053. | +/- | 105. |
| 2.76 | +/- | 0. |
| 2.43 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.72 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.43 |
| -9999.00 |

125-12
| 4113. | +/- | 1. |
| 4216. | +/- | 73. |
| 1.71 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.61 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.53 |
| -9999.00 |
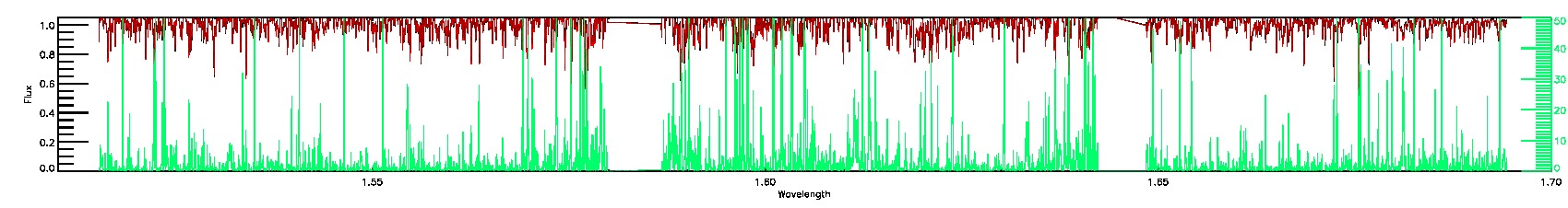
125-12
| 4875. | +/- | 12. |
| 4934. | +/- | 113. |
| 2.60 | +/- | 0. |
| 2.33 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.57 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.38 |
| -9999.00 |
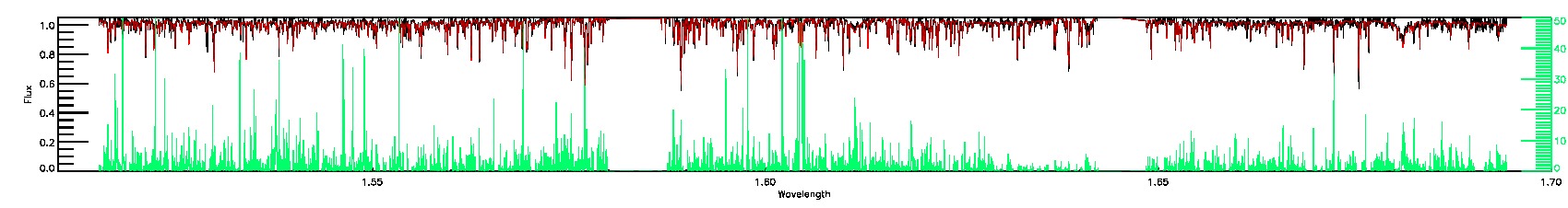
125-12
| 4367. | +/- | 5. |
| 4464. | +/- | 98. |
| 4.24 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 7.38 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.42 |
| -9999.00 |

125-12
| 4838. | +/- | 5. |
| 4891. | +/- | 88. |
| 2.88 | +/- | 0. |
| 2.51 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.13 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.85 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 8330. | +/- | 33. |
| 8157. | +/- | 269. |
| 4.16 | +/- | 0. |
| 4.02 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 71.14 | +/- | 0. |
| 1.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.75 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.46 |
| -9999.00 |
125-12
| 4969. | +/- | 8. |
| 4996. | +/- | 100. |
| 3.59 | +/- | 0. |
| 3.44 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |

125-12
| 4626. | +/- | 4. |
| 4707. | +/- | 83. |
| 2.65 | +/- | 0. |
| 2.45 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.60 |
| -9999.00 |

PERSIST_LOW
125-12
| 3982. | +/- | 1. |
| 4071. | +/- | 68. |
| 1.58 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.98 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
125-12
| 4848. | +/- | 5. |
| 4889. | +/- | 89. |
| 3.45 | +/- | 0. |
| 3.27 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.37 |
| -9999.00 |

125-12
| 4812. | +/- | 5. |
| 4871. | +/- | 88. |
| 2.91 | +/- | 0. |
| 2.73 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.39 |
| -9999.00 |

125-12
| 3723. | +/- | 1. |
| 3835. | +/- | 67. |
| 0.86 | +/- | 0. |
| 0.78 | +/- | 0. |
| 2.35 | +/- | 0. |
| 2.35 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -0.55 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.24 | +/- | 0. |
| 4.09 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.64 |
| -9999.00 |
STAR_BAD
STAR_WARN,COLORTE_WARN
125-12
| 8235. | +/- | 25. |
| -10000. | +/- | -NaN |
| 4.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 94.36 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.54 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.99 |
| -9999.00 |
125-12
| 4609. | +/- | 3. |
| 4681. | +/- | 80. |
| 2.65 | +/- | 0. |
| 2.35 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.80 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.09 |
| -9999.00 |

PERSIST_LOW
125-12
| 3746. | +/- | 1. |
| 3849. | +/- | 66. |
| 0.89 | +/- | 0. |
| 0.79 | +/- | 0. |
| 1.95 | +/- | 0. |
| 1.95 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.66 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.51 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
125-12
| 8000. | +/- | 12. |
| -10000. | +/- | -NaN |
| 4.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.29 | +/- | 1. |
| 1.29 | +/- | -NaN |
| -2.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.32 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.13 |
| -9999.00 |
125-12
| 4188. | +/- | 2. |
| 4303. | +/- | 84. |
| 1.72 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -0.62 | +/- | 0. |
| -0.62 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 4.25 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.38 |
| -9999.00 |
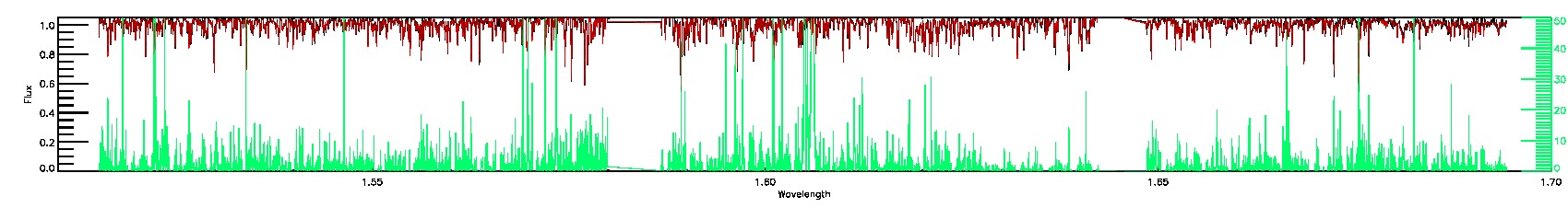
125-12
| 4850. | +/- | 5. |
| 4904. | +/- | 89. |
| 2.92 | +/- | 0. |
| 2.55 | +/- | 0. |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.20 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.36 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4873. | +/- | 6. |
| 4935. | +/- | 92. |
| 2.71 | +/- | 0. |
| 2.39 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.38 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.70 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.92 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4576. | +/- | 5. |
| 4657. | +/- | 89. |
| 2.56 | +/- | 0. |
| 2.31 | +/- | 0. |
| 1.23 | +/- | 0. |
| 1.23 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 3.05 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 8289. | +/- | 25. |
| 8114. | +/- | 260. |
| 4.37 | +/- | 0. |
| 4.21 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 39.73 | +/- | 0. |
| 1.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.21 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.18 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4900. | +/- | 8. |
| 4955. | +/- | 98. |
| 3.00 | +/- | 0. |
| 2.62 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.54 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.89 |
| -9999.00 |
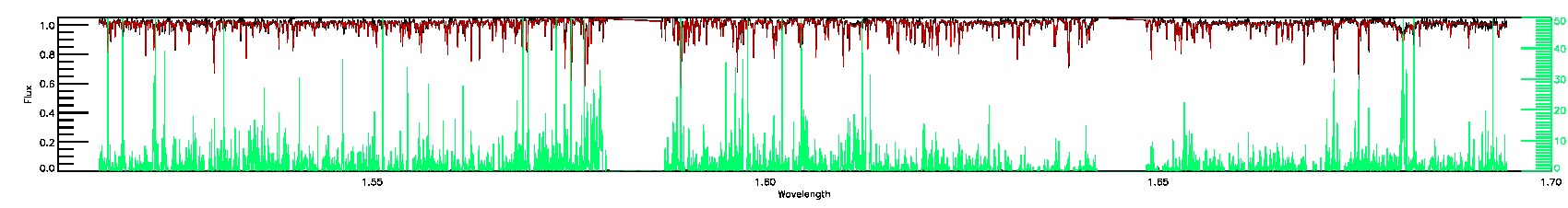
125-12
| 4952. | +/- | 14. |
| 5009. | +/- | 118. |
| 2.69 | +/- | 0. |
| 2.38 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -0.49 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 3.94 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.69 |
| -9999.00 |

125-12
| 4505. | +/- | 3. |
| 4614. | +/- | 83. |
| 2.25 | +/- | 0. |
| 2.08 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -0.49 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.93 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.65 |
| -9999.00 |

PERSIST_LOW
125-12
| 4160. | +/- | 1. |
| 4261. | +/- | 74. |
| 1.78 | +/- | 0. |
| 1.60 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.68 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.51 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.70 |
| -9999.00 |
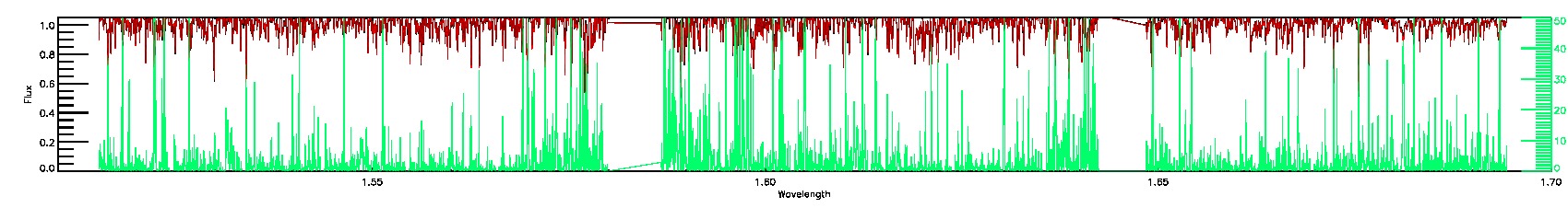
125-12
| 3979. | +/- | 1. |
| 4054. | +/- | 65. |
| 1.90 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.46 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.58 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
125-12
| 4538. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.52 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.06 |
| -9999.00 |
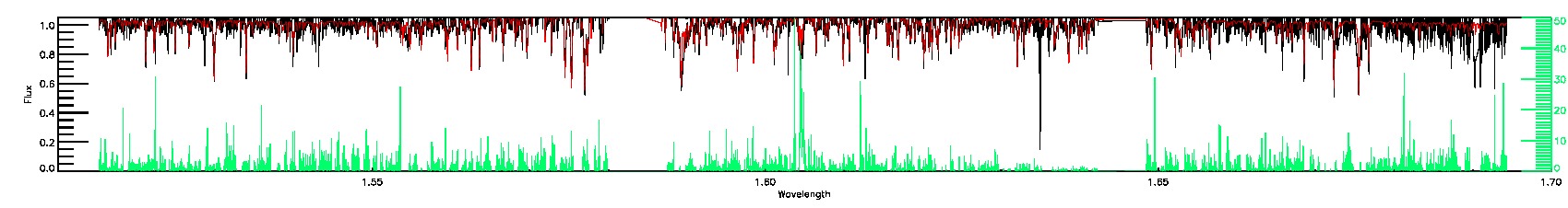
SUSPECT_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
125-12
| 8309. | +/- | 31. |
| -10000. | +/- | -NaN |
| 3.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 92.41 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.79 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.87 |
| -9999.00 |
125-12
| 4451. | +/- | 4. |
| 4536. | +/- | 91. |
| 4.38 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 6.22 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.22 |
| -9999.00 |
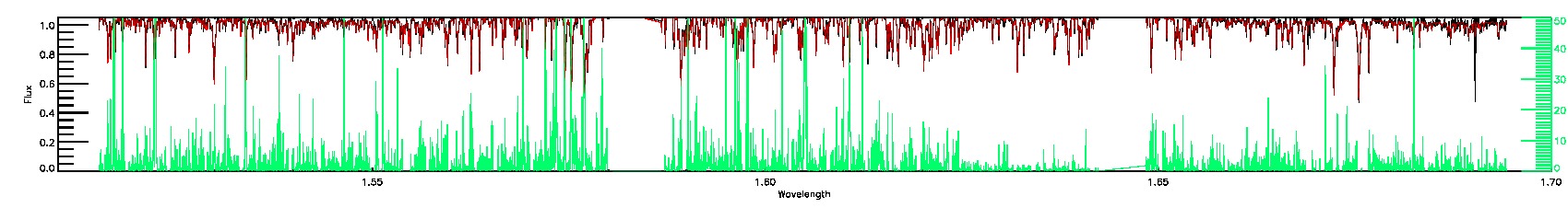
125-12
| 4662. | +/- | 7. |
| 4754. | +/- | 99. |
| 2.55 | +/- | 0. |
| 2.39 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -0.52 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.00 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.69 |
| -9999.00 |

125-12
| 4610. | +/- | 3. |
| 4687. | +/- | 82. |
| 2.64 | +/- | 0. |
| 2.35 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.04 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.36 |
| -9999.00 |

125-12
| 4976. | +/- | 7. |
| 5021. | +/- | 94. |
| 3.04 | +/- | 0. |
| 2.66 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.50 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
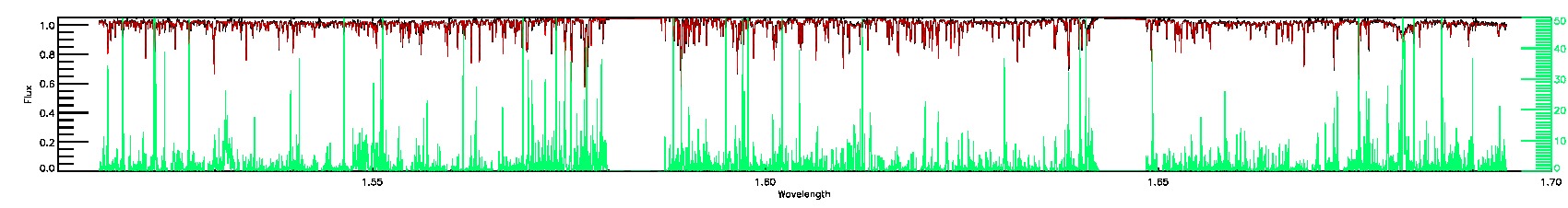
125-12
| 4849. | +/- | 6. |
| 4911. | +/- | 91. |
| 2.97 | +/- | 0. |
| 2.81 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.58 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.34 |
| -9999.00 |

125-12
| 4070. | +/- | 1. |
| 4179. | +/- | 73. |
| 1.50 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -0.49 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.93 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.90 |
| -9999.00 |

BRIGHT_NEIGHBOR
125-12
| 4733. | +/- | 6. |
| 4813. | +/- | 98. |
| 2.53 | +/- | 0. |
| 2.29 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.82 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.24 |
| -9999.00 |

SUSPECT_BROAD_LINES
125-12
| 3547. | +/- | 4. |
| 3642. | +/- | 73. |
| 4.46 | +/- | 0. |
| 4.85 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 19.38 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.54 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
125-12
| 8404. | +/- | 34. |
| 8247. | +/- | 283. |
| 4.38 | +/- | 0. |
| 4.38 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 27.31 | +/- | 0. |
| 1.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.04 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.22 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4805. | +/- | 5. |
| 4865. | +/- | 88. |
| 2.78 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.44 |
| -9999.00 |

125-12
| 3689. | +/- | 1. |
| 3805. | +/- | 67. |
| 0.58 | +/- | 0. |
| 0.52 | +/- | 0. |
| 2.27 | +/- | 0. |
| 2.27 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -0.65 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.89 |
| -9999.00 |
125-12
| 4950. | +/- | 12. |
| 4994. | +/- | 111. |
| 4.48 | +/- | 0. |
| 4.62 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.76 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 2.99 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.01 |
| -9999.00 |
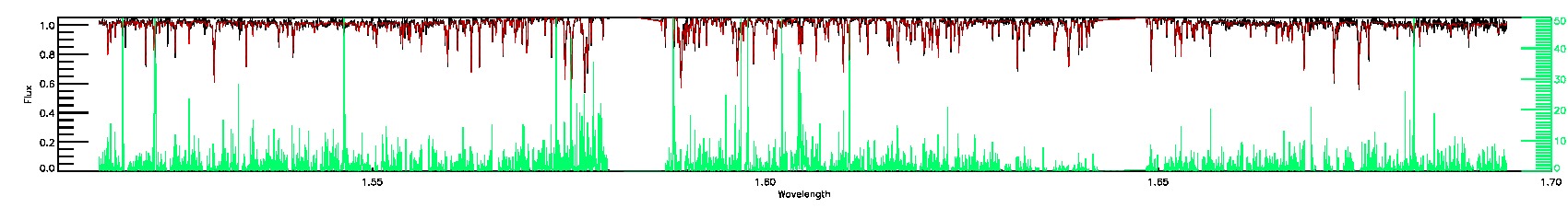
125-12
| 4081. | +/- | 1. |
| 4186. | +/- | 73. |
| 1.62 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.71 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.64 |
| -9999.00 |
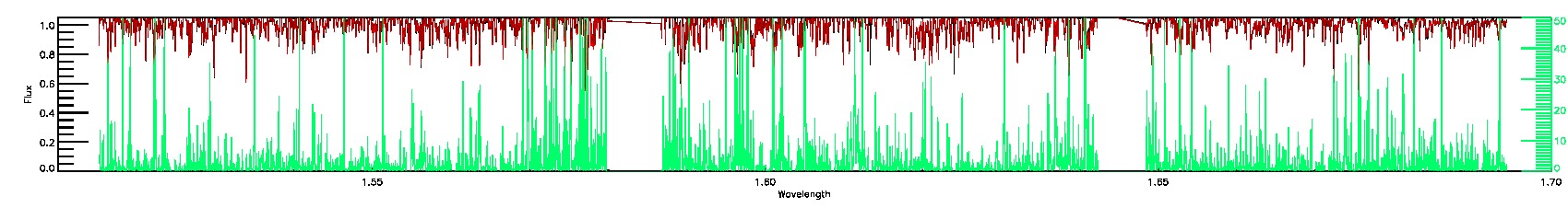
125-12
| 5182. | +/- | 14. |
| 5198. | +/- | 115. |
| 3.63 | +/- | 0. |
| 3.55 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.24 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.15 |
| -9999.00 |

125-12
| 4469. | +/- | 6. |
| 4583. | +/- | 105. |
| 2.00 | +/- | 0. |
| 1.84 | +/- | 0. |
| 1.60 | +/- | 0. |
| 1.60 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -0.60 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.21 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.54 |
| -9999.00 |

125-12
| 3956. | +/- | 1. |
| 4033. | +/- | 65. |
| 1.74 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.53 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.53 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
125-12
| 3537. | +/- | 1. |
| 3660. | +/- | 65. |
| 0.25 | +/- | 0. |
| 0.21 | +/- | 0. |
| 2.33 | +/- | 0. |
| 2.33 | +/- | -NaN |
| -0.81 | +/- | 0. |
| -0.81 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.33 |
| -9999.00 |
125-12
| 4317. | +/- | 2. |
| 4419. | +/- | 77. |
| 1.99 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.72 | +/- | 0. |
| 1.72 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.56 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.73 |
| -9999.00 |

125-12
| 4785. | +/- | 10. |
| 4847. | +/- | 106. |
| 2.88 | +/- | 0. |
| 2.70 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.25 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |

125-12
| 4224. | +/- | 1. |
| 4315. | +/- | 73. |
| 1.92 | +/- | 0. |
| 1.70 | +/- | 0. |
| 1.52 | +/- | 0. |
| 1.52 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.08 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.27 |
| -9999.00 |

125-12
| 4819. | +/- | 7. |
| 4889. | +/- | 102. |
| 2.43 | +/- | 0. |
| 2.25 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.68 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | 0. |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.80 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.22 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.71 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD
125-12
| 7997. | +/- | 14. |
| -10000. | +/- | -NaN |
| 4.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| -1.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 80.32 | +/- | 0. |
| 1.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.56 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 5151. | +/- | 17. |
| 5175. | +/- | 123. |
| 3.56 | +/- | 0. |
| 3.47 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.44 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |

125-12
| 4590. | +/- | 5. |
| 4676. | +/- | 95. |
| 2.53 | +/- | 0. |
| 2.29 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.53 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 3.29 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |

125-12
| 4687. | +/- | 8. |
| 4768. | +/- | 101. |
| 2.65 | +/- | 0. |
| 2.48 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.60 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.23 |
| -9999.00 |

125-12
| 3882. | +/- | 1. |
| 3989. | +/- | 69. |
| 1.16 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.86 | +/- | 0. |
| 1.86 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.78 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.94 |
| -9999.00 |
125-12
| 3751. | +/- | 1. |
| 3866. | +/- | 68. |
| 0.75 | +/- | 0. |
| 0.68 | +/- | 0. |
| 2.09 | +/- | 0. |
| 2.09 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -0.61 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.24 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.02 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4928. | +/- | 12. |
| 4988. | +/- | 112. |
| 2.71 | +/- | 0. |
| 2.39 | +/- | 0. |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -0.49 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.93 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.90 |
| -9999.00 |
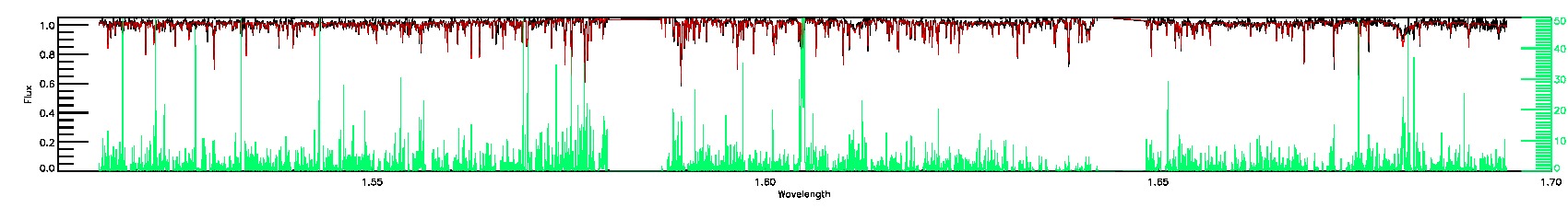
125-12
| 4436. | +/- | 2. |
| 4527. | +/- | 78. |
| 2.46 | +/- | 0. |
| 2.24 | +/- | 0. |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.06 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.34 |
| -9999.00 |
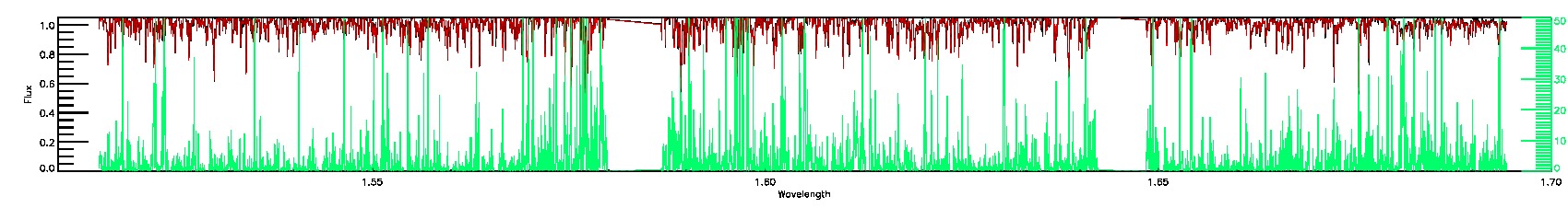
125-12
| 4645. | +/- | 7. |
| 4735. | +/- | 101. |
| 2.59 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.82 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.99 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 7937. | +/- | 16. |
| 7763. | +/- | 236. |
| 4.61 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 54.80 | +/- | 0. |
| 1.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.29 |
| -9999.00 |
125-12
| 4352. | +/- | 2. |
| 4449. | +/- | 77. |
| 2.13 | +/- | 0. |
| 1.92 | +/- | 0. |
| 1.55 | +/- | 0. |
| 1.55 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.35 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.76 |
| -9999.00 |

125-12
| 3802. | +/- | 1. |
| 3916. | +/- | 69. |
| 0.99 | +/- | 0. |
| 0.91 | +/- | 0. |
| 2.07 | +/- | 0. |
| 2.07 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -0.60 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.19 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.43 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
125-12
| 8068. | +/- | 23. |
| 7894. | +/- | 244. |
| 4.32 | +/- | 0. |
| 4.16 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 73.65 | +/- | 0. |
| 1.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.00 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.05 |
| -9999.00 |
125-12
| 4607. | +/- | 5. |
| 4678. | +/- | 94. |
| 2.91 | +/- | 0. |
| 2.69 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.78 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |

125-12
| 4546. | +/- | 4. |
| 4639. | +/- | 96. |
| 2.44 | +/- | 0. |
| 2.26 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.13 | +/- | 0. |
| 3.35 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.41 |
| -9999.00 |

125-12
| 4011. | +/- | 1. |
| 4119. | +/- | 72. |
| 1.44 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.89 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.79 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
125-12
| 8499. | +/- | 32. |
| 8384. | +/- | 327. |
| 3.96 | +/- | 0. |
| 4.37 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.43 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.35 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.20 |
| -9999.00 |
125-12
| 4155. | +/- | 1. |
| 4265. | +/- | 75. |
| 1.66 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.72 | +/- | 0. |
| 1.72 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.96 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.74 |
| -9999.00 |

SUSPECT_RV_COMBINATION
125-12
| 8301. | +/- | 25. |
| 8189. | +/- | 310. |
| 4.39 | +/- | 0. |
| 4.83 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.88 | +/- | 0. |
| 1.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.12 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.11 |
| -9999.00 |
125-12
| 4721. | +/- | 11. |
| 4787. | +/- | 111. |
| 2.98 | +/- | 0. |
| 2.80 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 3.09 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.27 |
| -9999.00 |

125-12
| 4429. | +/- | 2. |
| 4528. | +/- | 79. |
| 2.70 | +/- | 0. |
| 2.51 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.41 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.52 |
| -9999.00 |

125-12
| 3920. | +/- | 1. |
| 4030. | +/- | 70. |
| 1.30 | +/- | 0. |
| 1.21 | +/- | 0. |
| 1.99 | +/- | 0. |
| 1.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.20 | +/- | 0. |
| 3.95 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.91 |
| -9999.00 |
125-12
| 3740. | +/- | 1. |
| 3852. | +/- | 67. |
| 0.83 | +/- | 0. |
| 0.75 | +/- | 0. |
| 2.28 | +/- | 0. |
| 2.28 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -0.56 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.10 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.27 |
| -9999.00 |
125-12
| 4188. | +/- | 1. |
| 4270. | +/- | 70. |
| 2.09 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.70 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.15 |
| -9999.00 |
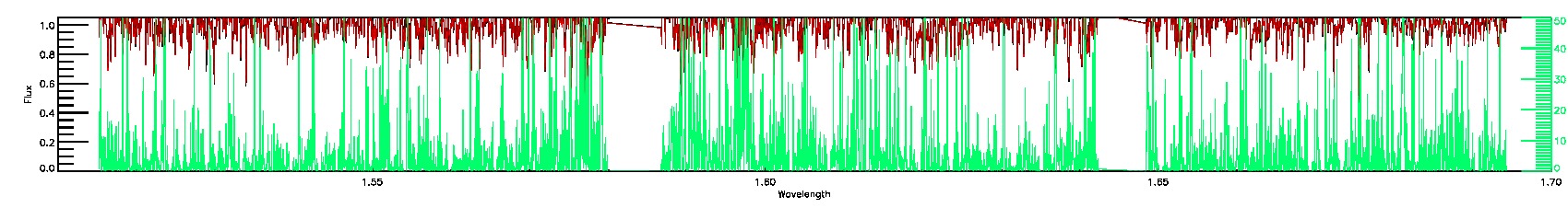
125-12
| 4746. | +/- | 7. |
| 4826. | +/- | 98. |
| 2.88 | +/- | 0. |
| 2.73 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.20 | +/- | 0. |
| 3.90 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.99 |
| -9999.00 |

125-12
| 3768. | +/- | 1. |
| 3855. | +/- | 63. |
| 1.36 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.62 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 2.92 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
125-12
| 4421. | +/- | 4. |
| 4533. | +/- | 92. |
| 2.09 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -0.55 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.09 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.19 |
| -9999.00 |

125-12
| 4553. | +/- | 4. |
| 4657. | +/- | 85. |
| 2.19 | +/- | 0. |
| 2.02 | +/- | 0. |
| 1.65 | +/- | 0. |
| 1.65 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 3.99 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.72 |
| -9999.00 |

125-12
| 3587. | +/- | 1. |
| 3700. | +/- | 64. |
| 0.43 | +/- | 0. |
| 0.35 | +/- | 0. |
| 2.27 | +/- | 0. |
| 2.27 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -0.57 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.14 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.79 |
| -9999.00 |
BRIGHT_NEIGHBOR
125-12
| 4005. | +/- | 1. |
| 4117. | +/- | 80. |
| 1.40 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.87 | +/- | 0. |
| 1.87 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -0.55 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.07 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.65 |
| -9999.00 |

125-12
| 4961. | +/- | 10. |
| 5012. | +/- | 121. |
| 2.73 | +/- | 0. |
| 2.41 | +/- | 0. |
| 1.60 | +/- | 0. |
| 1.60 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.67 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.95 |
| -9999.00 |

125-12
| 3879. | +/- | 1. |
| 3976. | +/- | 67. |
| 1.28 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.33 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
125-12
| 5046. | +/- | 18. |
| 5072. | +/- | 118. |
| 4.32 | +/- | 0. |
| 4.38 | +/- | 0. |
| 3.49 | +/- | 0. |
| 3.49 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 27.91 | +/- | 0. |
| 1.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 1.00 |
| -9999.00 |

125-12
| 4040. | +/- | 1. |
| 4133. | +/- | 70. |
| 1.71 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.17 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.23 |
| -9999.00 |

