SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 4816. | +/- | 10. |
| -10000. | +/- | -NaN |
| 1.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.73 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5399. | +/- | 14. |
| -10000. | +/- | -NaN |
| 3.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.33 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -2.33 |
| -9999.00 |

SUSPECT_RV_COMBINATION
135-03
| 8532. | +/- | 40. |
| 8418. | +/- | 330. |
| 4.30 | +/- | 0. |
| 4.72 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 55.94 | +/- | 0. |
| 1.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.29 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.04 |
| -9999.00 |
BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4386. | +/- | 30. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -1.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.56 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.76 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6081. | +/- | 27. |
| -10000. | +/- | -NaN |
| 3.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.12 | +/- | 0. |
| 1.12 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.76 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6182. | +/- | 26. |
| -10000. | +/- | -NaN |
| 3.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.52 | +/- | 1. |
| 1.52 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.45 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6404. | +/- | 29. |
| -10000. | +/- | -NaN |
| 4.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.83 | +/- | 1. |
| 1.83 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.08 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.38 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7453. | +/- | 15. |
| -10000. | +/- | -NaN |
| 4.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| -1.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.98 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.32 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5478. | +/- | 15. |
| -10000. | +/- | -NaN |
| 3.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.89 | +/- | 1. |
| 1.89 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6107. | +/- | 27. |
| -10000. | +/- | -NaN |
| 3.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.51 | +/- | 1. |
| 1.51 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.65 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.34 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4217. | +/- | 25. |
| -10000. | +/- | -NaN |
| 4.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.73 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.84 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6199. | +/- | 24. |
| -10000. | +/- | -NaN |
| 3.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5674. | +/- | 28. |
| -10000. | +/- | -NaN |
| 3.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.86 | +/- | 0. |
| 0.86 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.43 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.45 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 6937. | +/- | 4. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.48 | +/- | 0. |
| 4.48 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.97 | +/- | 0. |
| 1.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.52 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7998. | +/- | 12. |
| -10000. | +/- | -NaN |
| 4.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.87 | +/- | 1. |
| 0.87 | +/- | -NaN |
| -2.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.51 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.38 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5166. | +/- | 11. |
| -10000. | +/- | -NaN |
| 2.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5378. | +/- | 14. |
| -10000. | +/- | -NaN |
| 2.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.74 | +/- | 0. |
| 0.74 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6603. | +/- | 33. |
| -10000. | +/- | -NaN |
| 4.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 | +/- | 1. |
| 0.99 | +/- | -NaN |
| -2.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.74 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -2.29 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5465. | +/- | 13. |
| -10000. | +/- | -NaN |
| 2.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.03 | +/- | 0. |
| 2.03 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4344. | +/- | 44. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.64 | +/- | 1. |
| 0.64 | +/- | -NaN |
| -2.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.78 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.55 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.68 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 17846. | +/- | 196. |
| -10000. | +/- | -NaN |
| 4.48 | +/- | 0. |
| 3.90 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 78.23 | +/- | 0. |
| 1.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.52 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.56 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5912. | +/- | 20. |
| -10000. | +/- | -NaN |
| 3.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.74 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.52 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 8541. | +/- | 33. |
| 8412. | +/- | 318. |
| 4.02 | +/- | 0. |
| 4.29 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.99 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.99 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.50 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6190. | +/- | 29. |
| -10000. | +/- | -NaN |
| 3.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.27 | +/- | 1. |
| 1.27 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.88 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -2.49 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5633. | +/- | 16. |
| -10000. | +/- | -NaN |
| 3.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.20 | +/- | 1. |
| 1.20 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.18 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 12410. | +/- | 97. |
| -10000. | +/- | -NaN |
| 4.58 | +/- | 0. |
| 4.42 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.86 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.81 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.70 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6242. | +/- | 27. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.98 | +/- | 0. |
| 1.98 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.62 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.95 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6420. | +/- | 28. |
| -10000. | +/- | -NaN |
| 4.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.51 | +/- | 1. |
| 2.51 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.50 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.41 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5249. | +/- | 14. |
| -10000. | +/- | -NaN |
| 2.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.49 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
135-03
| 8889. | +/- | 56. |
| 8776. | +/- | 369. |
| 4.11 | +/- | 0. |
| 4.55 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 80.89 | +/- | 0. |
| 1.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.24 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.76 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 13850. | +/- | 102. |
| -10000. | +/- | -NaN |
| 4.53 | +/- | 0. |
| 4.23 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.40 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.29 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5542. | +/- | 13. |
| -10000. | +/- | -NaN |
| 3.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.69 | +/- | 0. |
| 4.69 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 14.83 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6146. | +/- | 29. |
| -10000. | +/- | -NaN |
| 3.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.33 | +/- | 2. |
| 1.33 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.30 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 4557. | +/- | 8. |
| -10000. | +/- | -NaN |
| 0.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.34 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6100. | +/- | 30. |
| -10000. | +/- | -NaN |
| 4.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.38 | +/- | 1. |
| 2.38 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.10 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.50 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 7926. | +/- | 20. |
| -10000. | +/- | -NaN |
| 5.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.85 | +/- | 1. |
| 1.85 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.69 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.35 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION
135-03
| 7719. | +/- | 16. |
| 7621. | +/- | 271. |
| 5.10 | +/- | 0. |
| 5.67 | +/- | 0. |
| 2.26 | +/- | 0. |
| 2.26 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -0.01 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.12 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.41 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 17068. | +/- | 200. |
| -10000. | +/- | -NaN |
| 4.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.37 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.77 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.77 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 5790. | +/- | 6. |
| -10000. | +/- | -NaN |
| 1.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.68 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.42 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 8115. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.46 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.58 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.55 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 8457. | +/- | 34. |
| -10000. | +/- | -NaN |
| 4.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 91.41 | +/- | 0. |
| 1.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.94 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.75 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6500. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.66 | +/- | 1. |
| 1.66 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.72 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.32 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4828. | +/- | 3. |
| -10000. | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.55 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION
135-03
| 7556. | +/- | 13. |
| 7458. | +/- | 258. |
| 4.74 | +/- | 0. |
| 5.31 | +/- | 0. |
| 0.78 | +/- | 2. |
| 0.78 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.09 | +/- | 3. |
| 0.09 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.14 | +/- | 1. |
| 0.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.43 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 6251. | +/- | 27. |
| -10000. | +/- | -NaN |
| 4.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.06 | +/- | 1. |
| 1.06 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.95 | +/- | 1. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -2.41 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5337. | +/- | 14. |
| -10000. | +/- | -NaN |
| 2.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 1. |
| 0.97 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.49 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6060. | +/- | 31. |
| -10000. | +/- | -NaN |
| 4.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.93 | +/- | 0. |
| 3.93 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.45 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5644. | +/- | 22. |
| -10000. | +/- | -NaN |
| 3.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.11 | +/- | 0. |
| 2.11 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.10 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 6111. | +/- | 27. |
| -10000. | +/- | -NaN |
| 3.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.27 | +/- | 1. |
| 1.27 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.69 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6147. | +/- | 24. |
| -10000. | +/- | -NaN |
| 3.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.89 | +/- | 1. |
| 0.89 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.77 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -2.28 |
| -9999.00 |
PERSIST_MED,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 7964. | +/- | 18. |
| -10000. | +/- | -NaN |
| 5.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.68 | +/- | 0. |
| 3.68 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.20 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.18 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6038. | +/- | 31. |
| -10000. | +/- | -NaN |
| 3.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.88 | +/- | 0. |
| 1.88 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.56 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.42 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6351. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.55 | +/- | 0. |
| 2.55 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.73 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5872. | +/- | 38. |
| -10000. | +/- | -NaN |
| 3.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.05 | +/- | 2. |
| 2.05 | +/- | -NaN |
| -2.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 92.30 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.94 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
135-03
| 8790. | +/- | 43. |
| 8675. | +/- | 356. |
| 4.22 | +/- | 0. |
| 4.63 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 22.94 | +/- | 0. |
| 1.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.19 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 7719. | +/- | 15. |
| -10000. | +/- | -NaN |
| 4.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.93 | +/- | 0. |
| 0.93 | +/- | -NaN |
| -1.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 90.09 | +/- | 0. |
| 1.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.52 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6128. | +/- | 27. |
| -10000. | +/- | -NaN |
| 3.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.14 | +/- | 1. |
| 1.14 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.71 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.43 |
| -9999.00 |
SUSPECT_RV_COMBINATION
135-03
| 7900. | +/- | 16. |
| 7778. | +/- | 269. |
| 4.26 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| -1.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -0.01 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 86.64 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.33 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6180. | +/- | 24. |
| -10000. | +/- | -NaN |
| 3.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.22 | +/- | 1. |
| 1.22 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.35 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 7994. | +/- | 13. |
| -10000. | +/- | -NaN |
| 4.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.75 | +/- | 1. |
| 0.75 | +/- | -NaN |
| -2.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.85 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.43 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7071. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.72 | +/- | 0. |
| 1.72 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.87 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 8000. | +/- | 15. |
| -10000. | +/- | -NaN |
| 5.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.46 | +/- | 0. |
| 4.46 | +/- | -NaN |
| -2.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -2.46 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7609. | +/- | 19. |
| -10000. | +/- | -NaN |
| 4.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.22 | +/- | 3. |
| 1.22 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.97 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 8635. | +/- | 89. |
| 8529. | +/- | 456. |
| 4.57 | +/- | 0. |
| 5.07 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.94 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.23 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.01 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6197. | +/- | 26. |
| -10000. | +/- | -NaN |
| 3.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.51 | +/- | 0. |
| 4.51 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.98 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5622. | +/- | 56. |
| -10000. | +/- | -NaN |
| 4.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.84 | +/- | 0. |
| 2.84 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.24 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
LOW_SNR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 4722. | +/- | 5. |
| -10000. | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.45 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7999. | +/- | 18. |
| -10000. | +/- | -NaN |
| 5.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.24 | +/- | 1. |
| 1.24 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.71 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5865. | +/- | 29. |
| -10000. | +/- | -NaN |
| 3.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.37 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.56 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 13893. | +/- | 150. |
| -10000. | +/- | -NaN |
| 4.46 | +/- | 0. |
| 4.40 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.60 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.68 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.66 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6908. | +/- | 22. |
| -10000. | +/- | -NaN |
| 4.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.80 | +/- | 1. |
| 1.80 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.22 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.86 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.47 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
135-03
| 8492. | +/- | 68. |
| 8381. | +/- | 405. |
| 4.49 | +/- | 0. |
| 4.95 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.49 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.97 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.94 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
135-03
| 13464. | +/- | 171. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| 4.43 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.59 | +/- | 1. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.84 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.86 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,ROTATION_WARN
135-03
| 4862. | +/- | 3. |
| -10000. | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.84 | +/- | 0. |
| 2.84 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.71 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.03 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6207. | +/- | 30. |
| -10000. | +/- | -NaN |
| 3.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| -2.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.85 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -2.27 |
| -9999.00 |
BRIGHT_NEIGHBOR
135-03
| 5954. | +/- | 15. |
| 5871. | +/- | 131. |
| 4.65 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 10.57 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.09 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6348. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.14 | +/- | 1. |
| 1.14 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.01 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.34 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7814. | +/- | 23. |
| -10000. | +/- | -NaN |
| 4.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 76.59 | +/- | 0. |
| 1.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -0.68 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,ROTATION_WARN
135-03
| 5876. | +/- | 5. |
| -10000. | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -1.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.01 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.06 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 14019. | +/- | 111. |
| -10000. | +/- | -NaN |
| 4.65 | +/- | 0. |
| 4.18 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 41.62 | +/- | 0. |
| 1.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.39 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6716. | +/- | 23. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.91 | +/- | 0. |
| 2.91 | +/- | -NaN |
| -2.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.63 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.43 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4989. | +/- | 20. |
| -10000. | +/- | -NaN |
| 2.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.74 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 12801. | +/- | 151. |
| -10000. | +/- | -NaN |
| 4.69 | +/- | 0. |
| 4.54 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.40 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.65 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.77 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 17094. | +/- | 192. |
| -10000. | +/- | -NaN |
| 4.58 | +/- | 0. |
| 4.09 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 39.29 | +/- | 0. |
| 1.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.33 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7592. | +/- | 18. |
| -10000. | +/- | -NaN |
| 4.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 2. |
| 0.83 | +/- | -NaN |
| -2.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.63 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.21 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5119. | +/- | 12. |
| -10000. | +/- | -NaN |
| 2.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -2.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.58 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.46 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 6197. | +/- | 26. |
| -10000. | +/- | -NaN |
| 3.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.14 | +/- | 1. |
| 1.14 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.24 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.32 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5871. | +/- | 23. |
| -10000. | +/- | -NaN |
| 3.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.58 | +/- | 1. |
| 1.58 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.50 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5489. | +/- | 17. |
| -10000. | +/- | -NaN |
| 3.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.35 | +/- | 1. |
| 1.35 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.46 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 5634. | +/- | 16. |
| -10000. | +/- | -NaN |
| 3.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.70 | +/- | 0. |
| 1.70 | +/- | -NaN |
| -2.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.30 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.64 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6166. | +/- | 30. |
| -10000. | +/- | -NaN |
| 3.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.53 | +/- | 1. |
| 1.53 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.03 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6528. | +/- | 23. |
| -10000. | +/- | -NaN |
| 4.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.72 | +/- | 1. |
| 1.72 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.61 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7119. | +/- | 27. |
| -10000. | +/- | -NaN |
| 4.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.91 | +/- | 2. |
| 1.91 | +/- | -NaN |
| -2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 93.78 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.37 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5962. | +/- | 28. |
| -10000. | +/- | -NaN |
| 3.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.23 | +/- | 0. |
| 2.23 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.96 |
| -9999.00 |

PERSIST_LOW,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5142. | +/- | 6. |
| -10000. | +/- | -NaN |
| 0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.58 | +/- | 0. |
| 0.58 | +/- | -NaN |
| -1.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.80 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6421. | +/- | 32. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.60 | +/- | 0. |
| 1.60 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.32 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
135-03
| 8603. | +/- | 34. |
| 8502. | +/- | 350. |
| 3.94 | +/- | 0. |
| 4.49 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.57 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.35 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.18 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6933. | +/- | 22. |
| -10000. | +/- | -NaN |
| 4.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.69 | +/- | 0. |
| 3.69 | +/- | -NaN |
| -2.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.87 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.37 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 12893. | +/- | 116. |
| -10000. | +/- | -NaN |
| 4.37 | +/- | 0. |
| 4.25 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.99 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.56 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6367. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.89 | +/- | 1. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.41 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,SN_WARN
135-03
| 6717. | +/- | 49. |
| 6558. | +/- | 217. |
| 4.33 | +/- | 0. |
| 4.15 | +/- | 0. |
| 0.56 | +/- | 0. |
| 0.56 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.17 | +/- | 0. |
| 28.97 | +/- | 0. |
| 1.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.26 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4215. | +/- | 25. |
| -10000. | +/- | -NaN |
| 4.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -2.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.31 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.97 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6101. | +/- | 20. |
| -10000. | +/- | -NaN |
| 3.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.06 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.44 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6459. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.28 | +/- | 0. |
| 3.28 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.77 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.49 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6597. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.23 | +/- | 1. |
| 1.23 | +/- | -NaN |
| -2.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.04 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.29 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
135-03
| 8516. | +/- | 37. |
| 8385. | +/- | 314. |
| 3.99 | +/- | 0. |
| 4.24 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 56.04 | +/- | 0. |
| 1.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.87 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.38 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6609. | +/- | 22. |
| -10000. | +/- | -NaN |
| 4.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.70 | +/- | 1. |
| 0.70 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.38 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.62 |
| -9999.00 |
PERSIST_LOW,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5118. | +/- | 4. |
| -10000. | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.68 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6141. | +/- | 27. |
| -10000. | +/- | -NaN |
| 3.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.00 | +/- | 0. |
| 3.00 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.93 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -2.20 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
135-03
| 7384. | +/- | 18. |
| 7286. | +/- | 246. |
| 5.08 | +/- | 0. |
| 5.68 | +/- | 0. |
| 3.80 | +/- | 0. |
| 3.80 | +/- | -NaN |
| -2.44 | +/- | 0. |
| -2.44 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.03 | +/- | 1. |
| -0.03 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.47 | +/- | 1. |
| 4.20 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.44 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 4950. | +/- | 8. |
| -10000. | +/- | -NaN |
| 1.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| -2.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.50 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.56 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
135-03
| 8246. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.02 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.61 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.11 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6331. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.45 | +/- | 0. |
| 4.45 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.36 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.34 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6606. | +/- | 23. |
| -10000. | +/- | -NaN |
| 4.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.92 | +/- | 1. |
| 1.92 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.03 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.45 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
135-03
| 8282. | +/- | 28. |
| 8158. | +/- | 298. |
| 4.13 | +/- | 0. |
| 4.44 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 74.61 | +/- | 0. |
| 1.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.72 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.31 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6286. | +/- | 18. |
| -10000. | +/- | -NaN |
| 3.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 1. |
| 0.83 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.91 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.35 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7998. | +/- | 34. |
| -10000. | +/- | -NaN |
| 5.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.16 | +/- | 4. |
| 1.16 | +/- | -NaN |
| -2.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.95 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -2.38 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4825. | +/- | 11. |
| -10000. | +/- | -NaN |
| 1.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.70 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6265. | +/- | 27. |
| -10000. | +/- | -NaN |
| 4.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.65 | +/- | 1. |
| 1.65 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.57 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.18 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5026. | +/- | 7. |
| -10000. | +/- | -NaN |
| 1.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| -1.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.08 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.25 |
| -9999.00 |

135-03
| 6329. | +/- | 17. |
| 6204. | +/- | 135. |
| 4.49 | +/- | 0. |
| 4.29 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 13.26 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4666. | +/- | 7. |
| -10000. | +/- | -NaN |
| 0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.70 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 7074. | +/- | 18. |
| -10000. | +/- | -NaN |
| 3.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| -1.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.92 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6206. | +/- | 27. |
| -10000. | +/- | -NaN |
| 4.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.21 | +/- | 0. |
| 2.21 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.58 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -2.34 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
135-03
| 7876. | +/- | 23. |
| 7778. | +/- | 284. |
| 5.34 | +/- | 0. |
| 5.91 | +/- | 0. |
| 2.21 | +/- | 1. |
| 2.21 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.36 | +/- | 1. |
| 0.36 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.16 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.25 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5603. | +/- | 22. |
| -10000. | +/- | -NaN |
| 3.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.01 | +/- | 1. |
| 1.01 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.42 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 7106. | +/- | 23. |
| -10000. | +/- | -NaN |
| 4.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.59 | +/- | 1. |
| 1.59 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.55 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.31 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 5210. | +/- | 8. |
| -10000. | +/- | -NaN |
| 1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.01 | +/- | 1. |
| 1.01 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

135-03
| 5062. | +/- | 6. |
| 5085. | +/- | 93. |
| 3.08 | +/- | 0. |
| 2.70 | +/- | 0. |
| 0.60 | +/- | 0. |
| 0.60 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.08 | +/- | 0. |
| 2.96 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
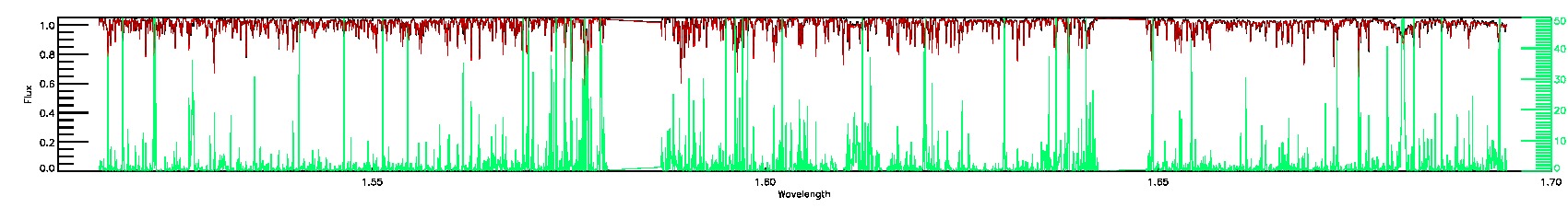
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5510. | +/- | 16. |
| -10000. | +/- | -NaN |
| 3.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.73 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.35 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.35 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6241. | +/- | 30. |
| -10000. | +/- | -NaN |
| 4.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.26 | +/- | 1. |
| 3.26 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.48 | +/- | 1. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.49 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4891. | +/- | 10. |
| -10000. | +/- | -NaN |
| 1.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.14 | +/- | 0. |
| 1.14 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.47 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6130. | +/- | 28. |
| -10000. | +/- | -NaN |
| 3.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.13 | +/- | 0. |
| 2.13 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.43 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 7422. | +/- | 19. |
| -10000. | +/- | -NaN |
| 5.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.52 | +/- | 1. |
| 1.52 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.90 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -2.28 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6277. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.67 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.41 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5641. | +/- | 15. |
| -10000. | +/- | -NaN |
| 3.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.19 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.42 |
| -9999.00 |
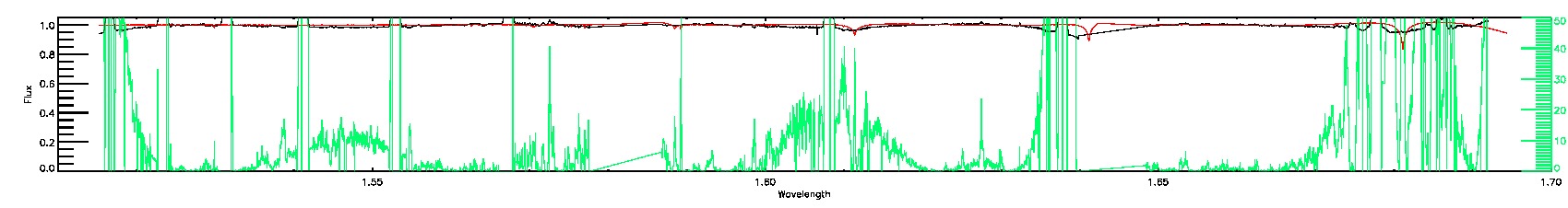
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5321. | +/- | 21. |
| -10000. | +/- | -NaN |
| 3.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.12 | +/- | 0. |
| 1.12 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.65 |
| -9999.00 |
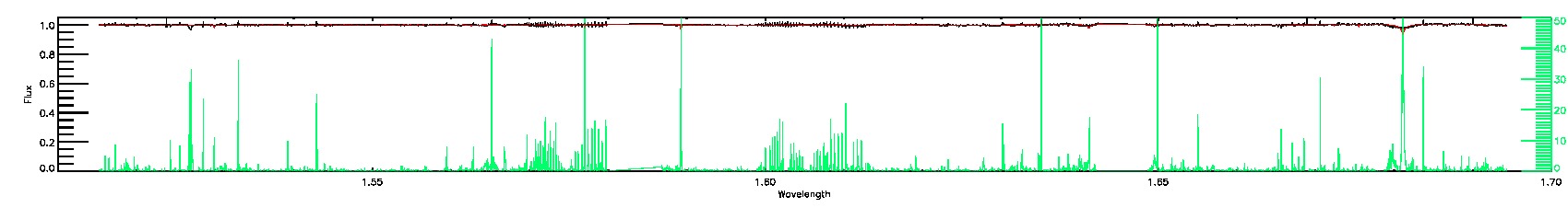
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 3941. | +/- | 8. |
| -10000. | +/- | -NaN |
| 2.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.28 | +/- | 0. |
| 2.28 | +/- | -NaN |
| -2.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.79 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7999. | +/- | 16. |
| -10000. | +/- | -NaN |
| 5.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.20 | +/- | 0. |
| 3.20 | +/- | -NaN |
| -2.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.53 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 16892. | +/- | 177. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| 4.09 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 31.75 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.02 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4770. | +/- | 52. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.71 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.18 |
| -9999.00 |
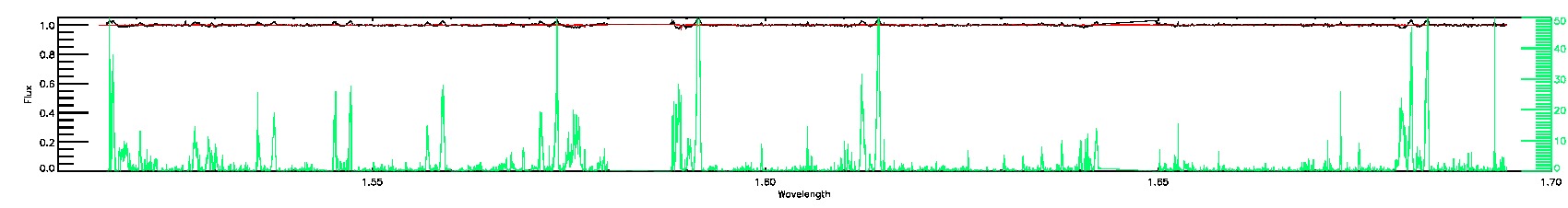
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 8711. | +/- | 54. |
| 8601. | +/- | 352. |
| 4.09 | +/- | 0. |
| 4.55 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 86.26 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.14 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.98 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6159. | +/- | 30. |
| -10000. | +/- | -NaN |
| 4.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.90 | +/- | 1. |
| 1.90 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.36 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.50 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 4119. | +/- | 11. |
| -10000. | +/- | -NaN |
| 3.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -1.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.83 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5935. | +/- | 44. |
| -10000. | +/- | -NaN |
| 4.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.91 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -2.49 |
| -9999.00 |
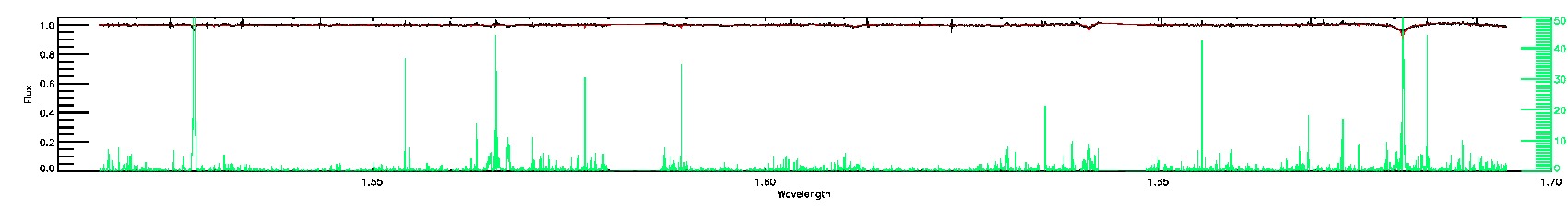
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4977. | +/- | 10. |
| -10000. | +/- | -NaN |
| 1.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.72 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.41 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 6514. | +/- | 27. |
| -10000. | +/- | -NaN |
| 3.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.04 | +/- | 0. |
| 2.04 | +/- | -NaN |
| -2.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.85 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.15 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6184. | +/- | 22. |
| -10000. | +/- | -NaN |
| 3.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.78 | +/- | 0. |
| 1.78 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.67 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.49 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6153. | +/- | 31. |
| -10000. | +/- | -NaN |
| 4.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.51 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -2.47 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 8660. | +/- | 51. |
| 8547. | +/- | 405. |
| 4.21 | +/- | 0. |
| 4.64 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.40 | +/- | 0. |
| 1.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.08 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.89 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5613. | +/- | 16. |
| -10000. | +/- | -NaN |
| 3.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.55 | +/- | 0. |
| 1.55 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
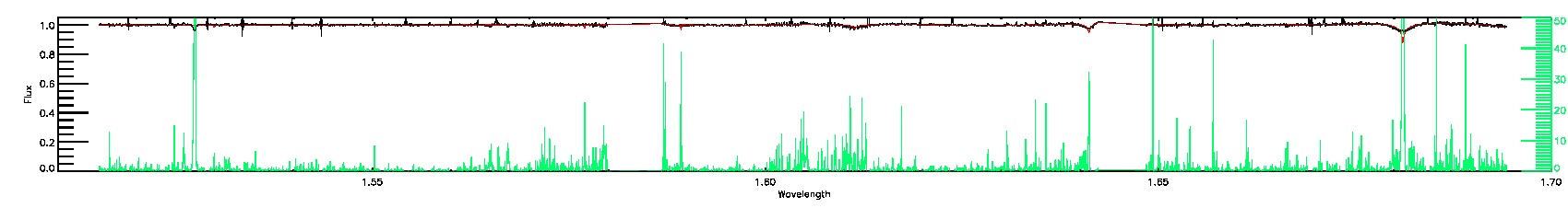
BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
135-03
| 14329. | +/- | 128. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| 4.07 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 31.94 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.36 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,SN_WARN
135-03
| 6740. | +/- | 69. |
| -10000. | +/- | -NaN |
| 4.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.57 | +/- | 1. |
| 1.57 | +/- | -NaN |
| -1.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.85 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.31 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 14361. | +/- | 123. |
| -10000. | +/- | -NaN |
| 4.54 | +/- | 0. |
| 4.13 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 22.55 | +/- | 0. |
| 1.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.20 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4801. | +/- | 12. |
| -10000. | +/- | -NaN |
| 2.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.59 | +/- | 0. |
| 3.59 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 6588. | +/- | 28. |
| -10000. | +/- | -NaN |
| 4.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.10 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5117. | +/- | 18. |
| -10000. | +/- | -NaN |
| 2.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.13 | +/- | 1. |
| 2.13 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 12458. | +/- | 68. |
| -10000. | +/- | -NaN |
| 4.42 | +/- | 0. |
| 4.15 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.32 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.38 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
135-03
| 7854. | +/- | 15. |
| 7755. | +/- | 282. |
| 4.94 | +/- | 0. |
| 5.50 | +/- | 0. |
| 0.87 | +/- | 1. |
| 0.87 | +/- | -NaN |
| -2.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.01 | +/- | 3. |
| 0.01 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.59 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.29 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 8470. | +/- | 29. |
| 8360. | +/- | 328. |
| 4.21 | +/- | 0. |
| 4.66 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.52 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.36 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -2.14 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6416. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.30 | +/- | 1. |
| 1.30 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.78 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.39 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6193. | +/- | 27. |
| -10000. | +/- | -NaN |
| 4.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.39 | +/- | 1. |
| 2.39 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.80 | +/- | 1. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.42 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6156. | +/- | 29. |
| -10000. | +/- | -NaN |
| 4.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.73 | +/- | 0. |
| 1.73 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.99 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 6256. | +/- | 28. |
| -10000. | +/- | -NaN |
| 4.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.54 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.50 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5264. | +/- | 8. |
| -10000. | +/- | -NaN |
| 1.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.75 | +/- | 0. |
| 0.75 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.53 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.47 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7995. | +/- | 42. |
| -10000. | +/- | -NaN |
| 5.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.03 | +/- | 1. |
| 1.03 | +/- | -NaN |
| -2.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.60 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.01 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6241. | +/- | 24. |
| -10000. | +/- | -NaN |
| 3.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.31 | +/- | 0. |
| 2.31 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.61 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.41 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4772. | +/- | 29. |
| -10000. | +/- | -NaN |
| 3.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.34 | +/- | 0. |
| 2.34 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.41 |
| -9999.00 |
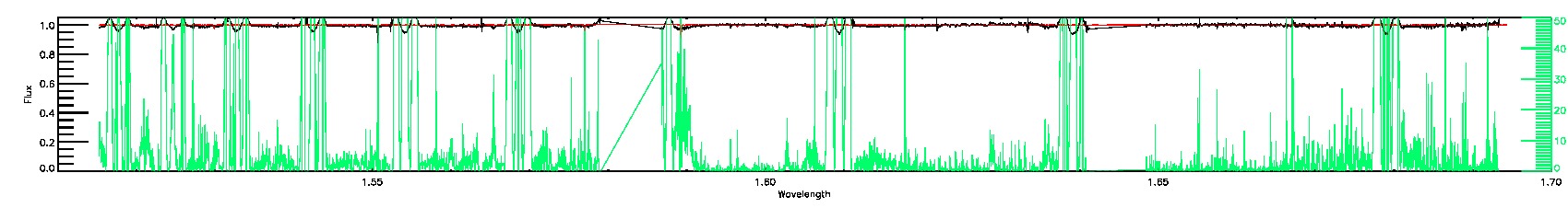
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6081. | +/- | 30. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 1. |
| 1.49 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.88 | +/- | 1. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.45 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 14444. | +/- | 122. |
| -10000. | +/- | -NaN |
| 4.56 | +/- | 0. |
| 4.28 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.22 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.20 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5456. | +/- | 10. |
| -10000. | +/- | -NaN |
| 2.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.70 | +/- | 0. |
| 1.70 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
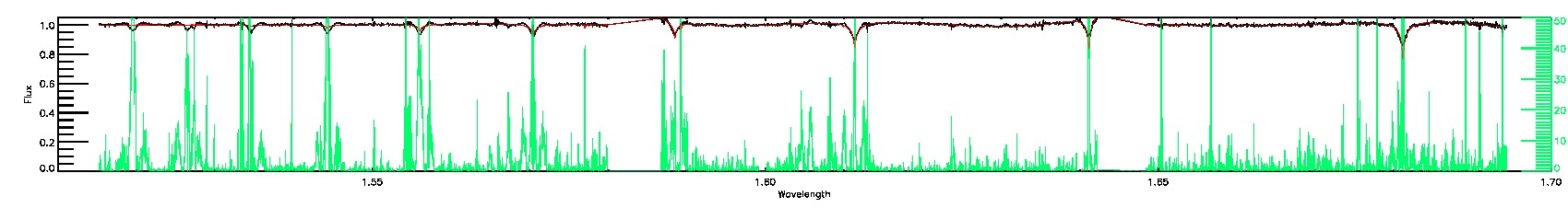
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 5090. | +/- | 17. |
| -10000. | +/- | -NaN |
| 2.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
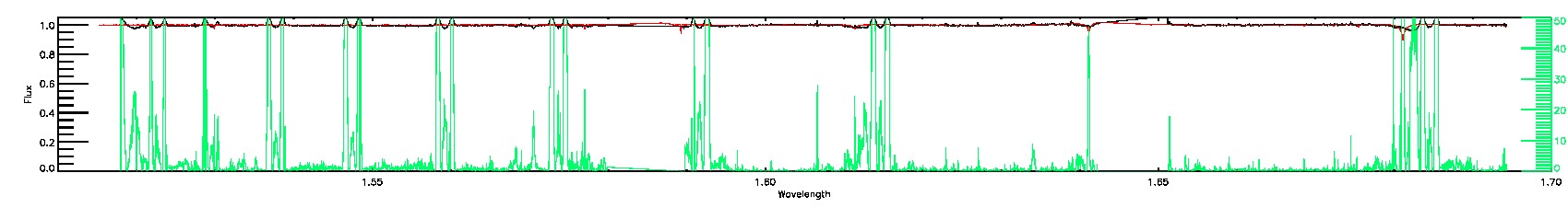
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 8867. | +/- | 69. |
| -10000. | +/- | -NaN |
| 3.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 90.32 | +/- | 0. |
| 1.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.26 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.83 |
| -9999.00 |
PERSIST_LOW,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 6094. | +/- | 17. |
| 6000. | +/- | 127. |
| 4.44 | +/- | 0. |
| 4.30 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 16.14 | +/- | 0. |
| 1.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5584. | +/- | 40. |
| -10000. | +/- | -NaN |
| 4.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.23 | +/- | 2. |
| 1.23 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
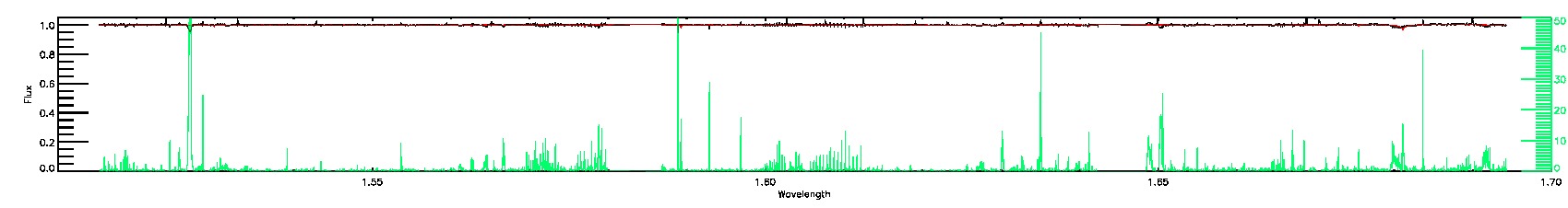
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6190. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.25 | +/- | 0. |
| 4.25 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.85 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.34 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 7587. | +/- | 17. |
| -10000. | +/- | -NaN |
| 5.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.89 | +/- | 1. |
| 1.89 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.37 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 8548. | +/- | 32. |
| -10000. | +/- | -NaN |
| 3.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.39 | +/- | 1. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.23 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 8731. | +/- | 44. |
| 8626. | +/- | 360. |
| 4.19 | +/- | 0. |
| 4.70 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.75 | +/- | 1. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.20 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.80 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5967. | +/- | 14. |
| -10000. | +/- | -NaN |
| 3.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.01 | +/- | 0. |
| 4.01 | +/- | -NaN |
| -2.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.05 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 7528. | +/- | 15. |
| -10000. | +/- | -NaN |
| 3.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.66 | +/- | 0. |
| 0.66 | +/- | -NaN |
| -1.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 94.49 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| 0.04 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 4288. | +/- | 7. |
| -10000. | +/- | -NaN |
| 0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.94 | +/- | 0. |
| 1.94 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.68 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.26 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 4710. | +/- | 21. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -2.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.47 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.37 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4856. | +/- | 12. |
| -10000. | +/- | -NaN |
| 2.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
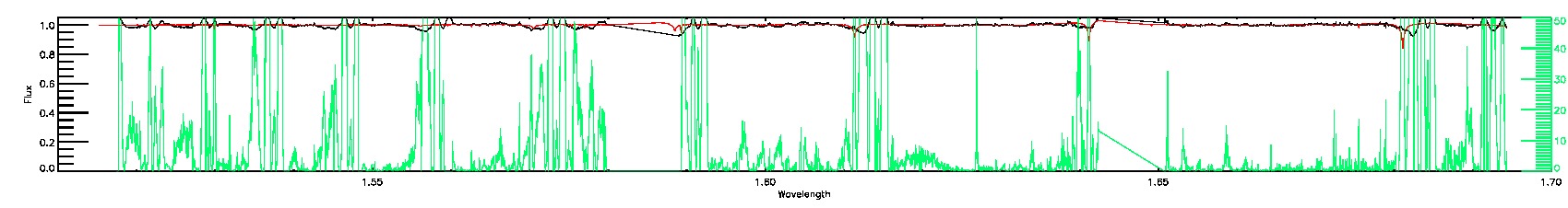
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5602. | +/- | 19. |
| -10000. | +/- | -NaN |
| 3.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.44 | +/- | 0. |
| 2.44 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.72 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5176. | +/- | 7. |
| -10000. | +/- | -NaN |
| 1.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.68 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5592. | +/- | 16. |
| -10000. | +/- | -NaN |
| 3.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.24 | +/- | 1. |
| 1.24 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.71 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.13 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 6630. | +/- | 22. |
| 6575. | +/- | 196. |
| 4.40 | +/- | 0. |
| 5.13 | +/- | 0. |
| 3.03 | +/- | 0. |
| 3.03 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -2.41 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| -0.15 | +/- | 1. |
| -0.15 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.48 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.43 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
135-03
| 8328. | +/- | 32. |
| 8208. | +/- | 307. |
| 4.25 | +/- | 0. |
| 4.62 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 80.97 | +/- | 0. |
| 1.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.45 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -1.96 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 8501. | +/- | 29. |
| 8399. | +/- | 339. |
| 4.27 | +/- | 0. |
| 4.81 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.33 | +/- | 0. |
| 1.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.32 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.20 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5902. | +/- | 36. |
| -10000. | +/- | -NaN |
| 3.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.47 | +/- | 0. |
| 2.47 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.58 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7998. | +/- | 18. |
| -10000. | +/- | -NaN |
| 4.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.92 | +/- | 0. |
| 1.92 | +/- | -NaN |
| -1.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 90.14 | +/- | 0. |
| 1.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.95 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5363. | +/- | 14. |
| -10000. | +/- | -NaN |
| 3.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -2.50 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION
135-03
| 8382. | +/- | 33. |
| 8281. | +/- | 328. |
| 4.12 | +/- | 0. |
| 4.67 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 42.73 | +/- | 0. |
| 1.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.22 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7360. | +/- | 18. |
| -10000. | +/- | -NaN |
| 4.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.66 | +/- | 3. |
| 0.66 | +/- | -NaN |
| -2.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.43 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -2.29 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6147. | +/- | 30. |
| -10000. | +/- | -NaN |
| 4.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.54 | +/- | 1. |
| 1.54 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.92 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -2.42 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6728. | +/- | 21. |
| -10000. | +/- | -NaN |
| 4.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.59 | +/- | 1. |
| 1.59 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.99 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 5329. | +/- | 17. |
| -10000. | +/- | -NaN |
| 3.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.41 |
| -9999.00 |
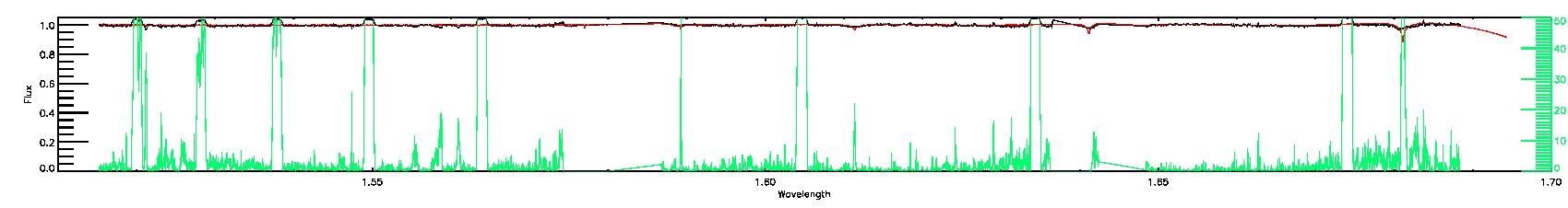
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6000. | +/- | 6. |
| -10000. | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| -1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.50 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.85 |
| -9999.00 |
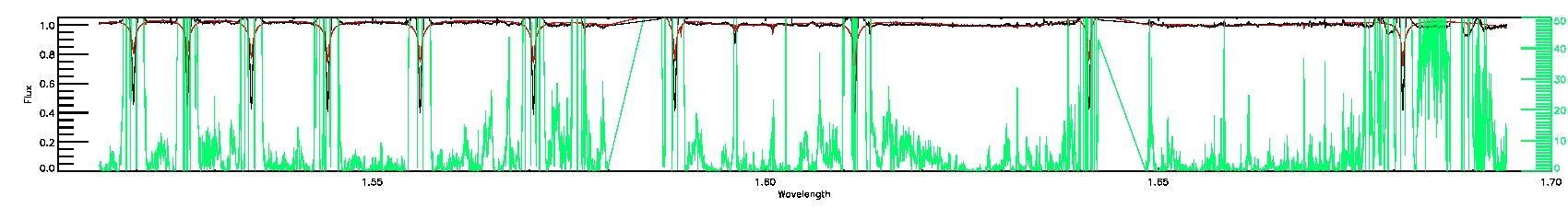
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4375. | +/- | 15. |
| -10000. | +/- | -NaN |
| 1.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 8146. | +/- | 22. |
| 7989. | +/- | 262. |
| 4.05 | +/- | 0. |
| 4.06 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 31.90 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.18 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.98 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6125. | +/- | 28. |
| -10000. | +/- | -NaN |
| 4.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.18 | +/- | 0. |
| 2.18 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.94 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5946. | +/- | 37. |
| -10000. | +/- | -NaN |
| 3.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.45 | +/- | 1. |
| 1.45 | +/- | -NaN |
| -2.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.98 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.28 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4520. | +/- | 8. |
| -10000. | +/- | -NaN |
| 0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.91 | +/- | 0. |
| 1.91 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.74 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.65 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.02 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5440. | +/- | 13. |
| -10000. | +/- | -NaN |
| 2.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.49 |
| -9999.00 |
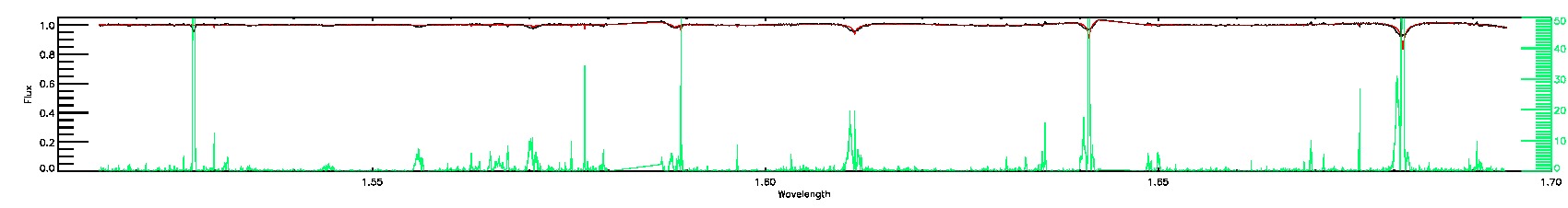
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6701. | +/- | 22. |
| -10000. | +/- | -NaN |
| 4.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.91 | +/- | 0. |
| 1.91 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.40 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -2.33 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_WARN,COLORTE_WARN
135-03
| 12661. | +/- | 66. |
| -10000. | +/- | -NaN |
| 4.41 | +/- | 0. |
| 4.14 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.59 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.35 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4935. | +/- | 14. |
| -10000. | +/- | -NaN |
| 2.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.72 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
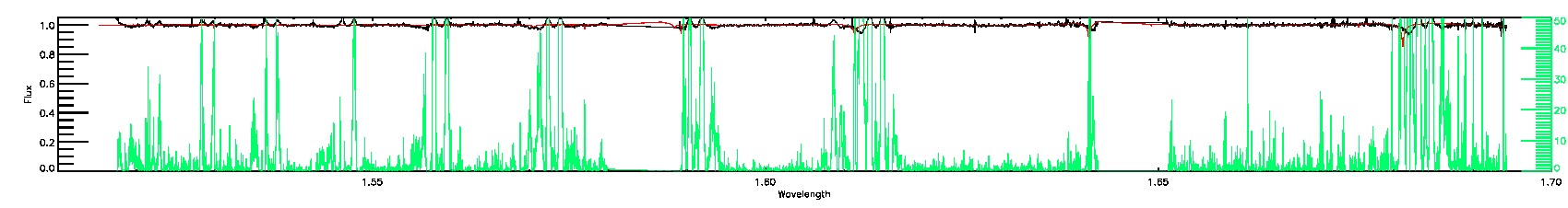
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6297. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.62 | +/- | 0. |
| 2.62 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.91 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4721. | +/- | 5. |
| -10000. | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.36 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 6334. | +/- | 22. |
| -10000. | +/- | -NaN |
| 3.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.35 | +/- | 0. |
| 2.35 | +/- | -NaN |
| -2.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.65 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4255. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.78 | +/- | 0. |
| 4.78 | +/- | -NaN |
| -2.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.99 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.22 |
| -9999.00 |
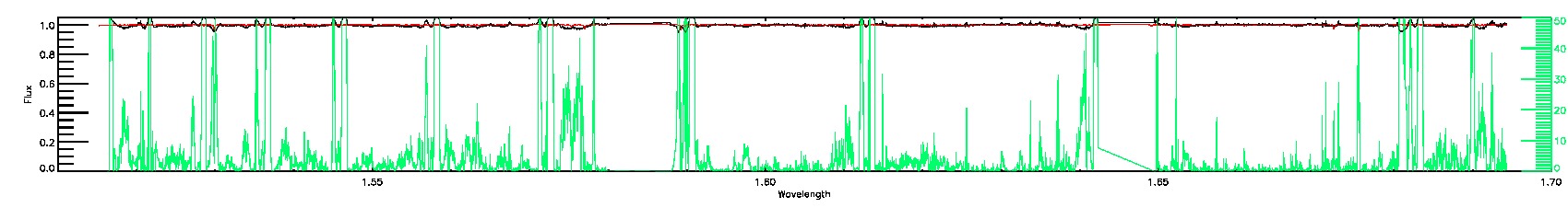
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6590. | +/- | 24. |
| -10000. | +/- | -NaN |
| 4.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.04 | +/- | 1. |
| 1.04 | +/- | -NaN |
| -2.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.60 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.40 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5126. | +/- | 21. |
| -10000. | +/- | -NaN |
| 3.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
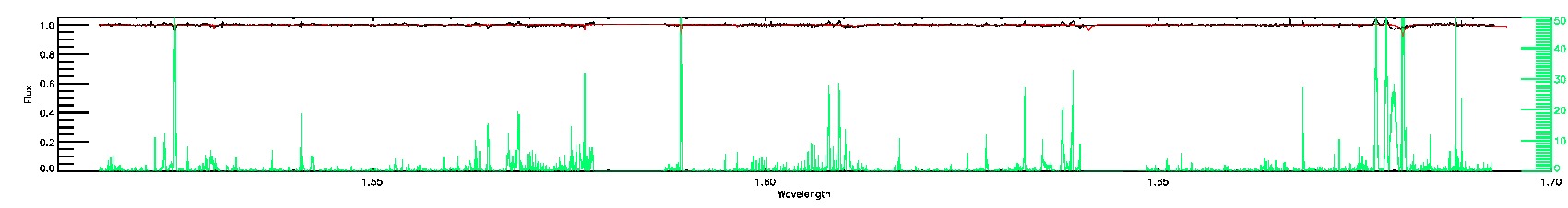
SUSPECT_RV_COMBINATION
STAR_BAD
135-03
| 7380. | +/- | 25. |
| -10000. | +/- | -NaN |
| 4.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.80 | +/- | 1. |
| 1.80 | +/- | -NaN |
| -2.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.72 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.47 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5762. | +/- | 39. |
| -10000. | +/- | -NaN |
| 3.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 91.54 | +/- | 0. |
| 1.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.47 |
| -9999.00 |
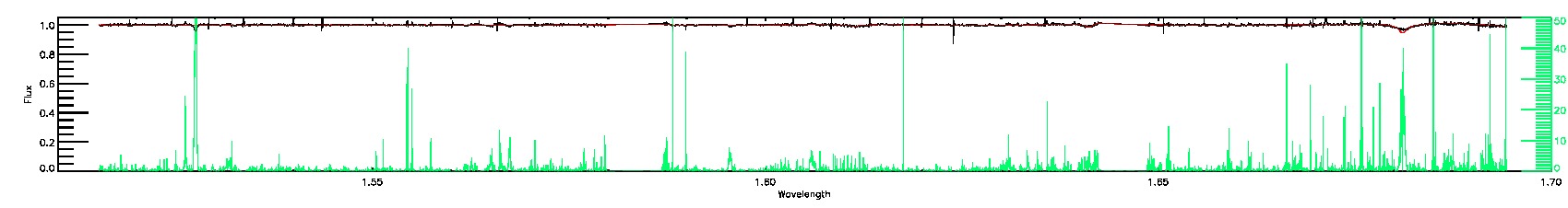
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 8869. | +/- | 49. |
| -10000. | +/- | -NaN |
| 3.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.81 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.58 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.03 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6250. | +/- | 23. |
| -10000. | +/- | -NaN |
| 3.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.47 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.36 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4782. | +/- | 16. |
| -10000. | +/- | -NaN |
| 2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.97 | +/- | 0. |
| 1.97 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.71 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6095. | +/- | 24. |
| -10000. | +/- | -NaN |
| 3.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.71 | +/- | 0. |
| 2.71 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.61 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4565. | +/- | 8. |
| -10000. | +/- | -NaN |
| 0.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.23 | +/- | 0. |
| 1.23 | +/- | -NaN |
| -2.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.04 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.41 |
| -9999.00 |

PERSIST_LOW,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 6204. | +/- | 28. |
| -10000. | +/- | -NaN |
| 4.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.74 | +/- | 0. |
| 4.74 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.40 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
135-03
| 8192. | +/- | 27. |
| -10000. | +/- | -NaN |
| 4.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 36.79 | +/- | 0. |
| 1.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
135-03
| 8395. | +/- | 26. |
| 8296. | +/- | 330. |
| 4.11 | +/- | 0. |
| 4.67 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.14 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.38 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.40 |
| -9999.00 |
PERSIST_LOW,SUSPECT_RV_COMBINATION
135-03
| 7958. | +/- | 22. |
| 7813. | +/- | 260. |
| 4.67 | +/- | 0. |
| 4.79 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.19 | +/- | -NaN |
| -1.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.40 | +/- | 1. |
| 0.40 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 84.00 | +/- | 0. |
| 1.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -1.32 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 8703. | +/- | 49. |
| -10000. | +/- | -NaN |
| 4.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 94.71 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.90 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.53 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4686. | +/- | 3. |
| -10000. | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.73 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.36 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6618. | +/- | 21. |
| -10000. | +/- | -NaN |
| 4.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.14 | +/- | 1. |
| 1.14 | +/- | -NaN |
| -2.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.55 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4759. | +/- | 23. |
| -10000. | +/- | -NaN |
| 2.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.77 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6618. | +/- | 23. |
| -10000. | +/- | -NaN |
| 4.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.70 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -2.47 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5173. | +/- | 16. |
| -10000. | +/- | -NaN |
| 2.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.86 | +/- | 0. |
| 3.86 | +/- | -NaN |
| -2.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.29 | +/- | 0. |
| 1.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.46 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7999. | +/- | 12. |
| -10000. | +/- | -NaN |
| 4.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.86 | +/- | 1. |
| 0.86 | +/- | -NaN |
| -2.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.72 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.05 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5763. | +/- | 16. |
| -10000. | +/- | -NaN |
| 3.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.34 | +/- | 1. |
| 1.34 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
135-03
| 8282. | +/- | 34. |
| 8174. | +/- | 312. |
| 4.24 | +/- | 0. |
| 4.72 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 56.77 | +/- | 0. |
| 1.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.94 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6458. | +/- | 22. |
| -10000. | +/- | -NaN |
| 4.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.91 | +/- | 1. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.40 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6091. | +/- | 25. |
| -10000. | +/- | -NaN |
| 3.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.78 | +/- | 1. |
| 1.78 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.54 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5294. | +/- | 12. |
| -10000. | +/- | -NaN |
| 2.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| -2.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.97 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.48 |
| -9999.00 |
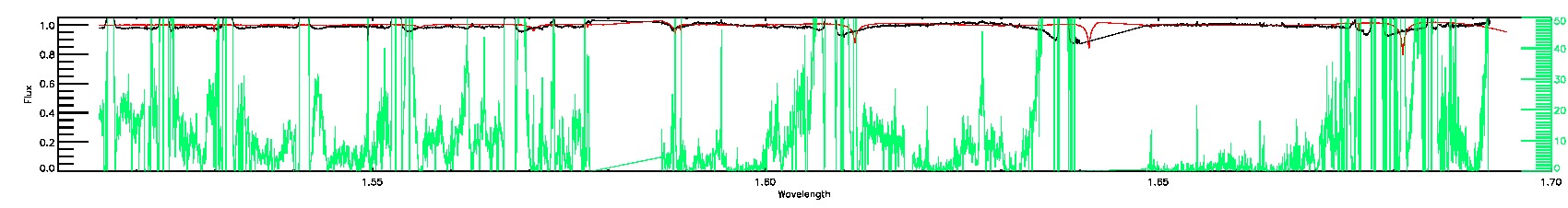
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6166. | +/- | 22. |
| -10000. | +/- | -NaN |
| 3.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 65.45 | +/- | 0. |
| 1.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -2.23 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7935. | +/- | 12. |
| -10000. | +/- | -NaN |
| 4.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.73 | +/- | 0. |
| 0.73 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 43.29 | +/- | 0. |
| 1.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.99 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN
135-03
| 5355. | +/- | 25. |
| -10000. | +/- | -NaN |
| 3.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.60 | +/- | 0. |
| 0.60 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.52 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.43 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5549. | +/- | 14. |
| -10000. | +/- | -NaN |
| 3.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.95 | +/- | 0. |
| 0.95 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.41 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.75 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -2.42 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5563. | +/- | 8. |
| -10000. | +/- | -NaN |
| 3.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.50 | +/- | 1. |
| 1.50 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.61 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.62 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6848. | +/- | 4. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| -1.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 86.22 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.40 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7910. | +/- | 16. |
| -10000. | +/- | -NaN |
| 5.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.29 | +/- | 1. |
| 4.29 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.62 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.44 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5668. | +/- | 14. |
| -10000. | +/- | -NaN |
| 3.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.34 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
135-03
| 8755. | +/- | 39. |
| 8638. | +/- | 350. |
| 3.94 | +/- | 0. |
| 4.33 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.64 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.14 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.96 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,CHI2_WARN,COLORTE_WARN
135-03
| 6504. | +/- | 4. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.39 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.13 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
135-03
| 7945. | +/- | 20. |
| -10000. | +/- | -NaN |
| 5.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.90 | +/- | 2. |
| 1.90 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.42 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.44 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6177. | +/- | 23. |
| -10000. | +/- | -NaN |
| 3.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.23 | +/- | 0. |
| 2.23 | +/- | -NaN |
| -2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.26 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.36 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5941. | +/- | 75. |
| -10000. | +/- | -NaN |
| 4.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.24 | +/- | 1. |
| 1.24 | +/- | -NaN |
| -2.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.00 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5942. | +/- | 29. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.56 | +/- | 1. |
| 1.56 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.72 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -2.49 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6391. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.56 | +/- | 0. |
| 4.56 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.14 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.44 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 4729. | +/- | 8. |
| -10000. | +/- | -NaN |
| 1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.74 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.22 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6193. | +/- | 30. |
| -10000. | +/- | -NaN |
| 4.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.87 | +/- | 1. |
| 1.87 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.18 | +/- | 1. |
| 0.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5830. | +/- | 40. |
| -10000. | +/- | -NaN |
| 3.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.05 | +/- | 1. |
| 1.05 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 92.00 | +/- | 0. |
| 1.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.18 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6319. | +/- | 20. |
| -10000. | +/- | -NaN |
| 3.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.34 | +/- | 1. |
| 1.34 | +/- | -NaN |
| -2.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.60 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5353. | +/- | 13. |
| -10000. | +/- | -NaN |
| 2.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5673. | +/- | 14. |
| -10000. | +/- | -NaN |
| 3.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 3987. | +/- | 21. |
| -10000. | +/- | -NaN |
| 4.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.52 | +/- | 0. |
| 1.52 | +/- | -NaN |
| -1.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.72 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.04 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5075. | +/- | 6. |
| -10000. | +/- | -NaN |
| 0.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.40 | +/- | 0. |
| 4.40 | +/- | -NaN |
| -2.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.67 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.42 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
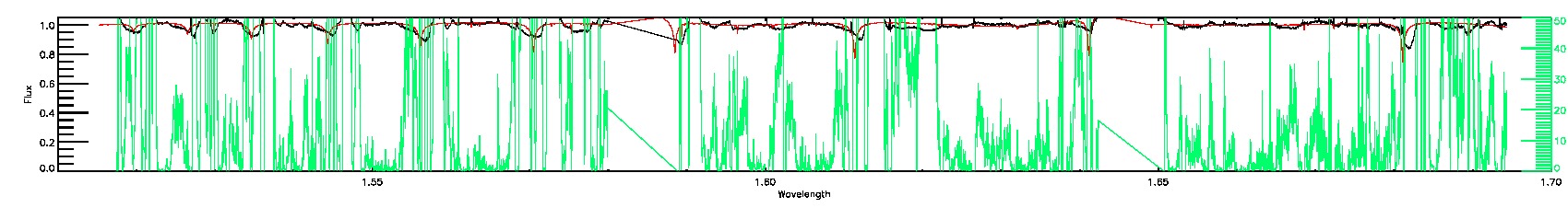
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6858. | +/- | 18. |
| -10000. | +/- | -NaN |
| 4.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.35 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.33 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 7376. | +/- | 13. |
| -10000. | +/- | -NaN |
| 4.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| -2.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.66 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.16 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
135-03
| 5410. | +/- | 15. |
| -10000. | +/- | -NaN |
| 3.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.78 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
135-03
| 5189. | +/- | 19. |
| -10000. | +/- | -NaN |
| 2.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.13 | +/- | 1. |
| 1.13 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.17 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
135-03
| 6085. | +/- | 28. |
| -10000. | +/- | -NaN |
| 3.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.59 | +/- | 1. |
| 1.59 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.92 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -2.47 |
| -9999.00 |
