170-60
| 5757. | +/- | 17. |
| 5701. | +/- | 126. |
| 4.37 | +/- | 0. |
| 4.30 | +/- | 0. |
| 0.70 | +/- | 0. |
| 0.70 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.59 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.47 |
| -9999.00 |
170-60
| 5653. | +/- | 20. |
| 5610. | +/- | 131. |
| 4.32 | +/- | 0. |
| 4.27 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.46 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW
STAR_WARN,SN_WARN
170-60
| 5765. | +/- | 38. |
| 5718. | +/- | 164. |
| 4.26 | +/- | 0. |
| 4.28 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.16 | +/- | 0. |
| 3.12 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.54 |
| -9999.00 |
BRIGHT_NEIGHBOR
170-60
| 3947. | +/- | 2. |
| 4040. | +/- | 76. |
| 4.41 | +/- | 0. |
| 4.72 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.20 |
| -9999.00 |
170-60
| 4210. | +/- | 2. |
| 4289. | +/- | 71. |
| 4.46 | +/- | 0. |
| 4.57 | +/- | 0. |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.08 | +/- | 0. |
| 1.72 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.36 |
| -9999.00 |
170-60
| 4525. | +/- | 4. |
| 4637. | +/- | 89. |
| 3.98 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| -0.63 | +/- | 0. |
| -0.62 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 6.61 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.35 |
| -9999.00 |
PERSIST_LOW
170-60
| 4875. | +/- | 5. |
| 4933. | +/- | 91. |
| 2.76 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.65 | +/- | 0. |
| 1.65 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.58 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.24 |
| -9999.00 |

170-60
| 5744. | +/- | 18. |
| 5683. | +/- | 132. |
| 4.30 | +/- | 0. |
| 4.16 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 4.12 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5462. | +/- | 14. |
| 5445. | +/- | 123. |
| 4.02 | +/- | 0. |
| 4.05 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 3.23 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
PERSIST_LOW
170-60
| 4847. | +/- | 10. |
| 4918. | +/- | 114. |
| 2.84 | +/- | 0. |
| 2.48 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -0.54 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 4.06 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
170-60
| 4032. | +/- | 2. |
| 4125. | +/- | 69. |
| 4.42 | +/- | 0. |
| 4.70 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
170-60
| 5943. | +/- | 14. |
| 5881. | +/- | 126. |
| 4.00 | +/- | 0. |
| 4.04 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 7.58 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
PERSIST_LOW,SUSPECT_BROAD_LINES
170-60
| 5435. | +/- | 13. |
| 5419. | +/- | 105. |
| 4.24 | +/- | 0. |
| 4.24 | +/- | 0. |
| 2.02 | +/- | 0. |
| 2.02 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 24.43 | +/- | 0. |
| 1.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |

PERSIST_LOW
170-60
| 4831. | +/- | 6. |
| 4900. | +/- | 98. |
| 4.06 | +/- | 0. |
| 4.33 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.19 | +/- | 0. |
| 6.05 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.23 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
170-60
| 5882. | +/- | 8. |
| 5816. | +/- | 120. |
| 4.10 | +/- | 0. |
| 4.04 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 45.31 | +/- | 0. |
| 1.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.08 |
| -9999.00 |
170-60
| 6505. | +/- | 20. |
| 6368. | +/- | 146. |
| 4.40 | +/- | 0. |
| 4.24 | +/- | 0. |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 13.61 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.40 |
| -9999.00 |
170-60
| 4937. | +/- | 8. |
| 5001. | +/- | 101. |
| 2.70 | +/- | 0. |
| 2.38 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -0.60 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.25 | +/- | 0. |
| 4.21 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.45 |
| -9999.00 |

PERSIST_LOW
170-60
| 4708. | +/- | 6. |
| 4768. | +/- | 99. |
| 4.55 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 6.44 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
170-60
| 5487. | +/- | 9. |
| 5457. | +/- | 104. |
| 4.59 | +/- | 0. |
| 4.52 | +/- | 0. |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.31 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.36 |
| -9999.00 |

PERSIST_LOW
170-60
| 5386. | +/- | 9. |
| 5373. | +/- | 103. |
| 4.62 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
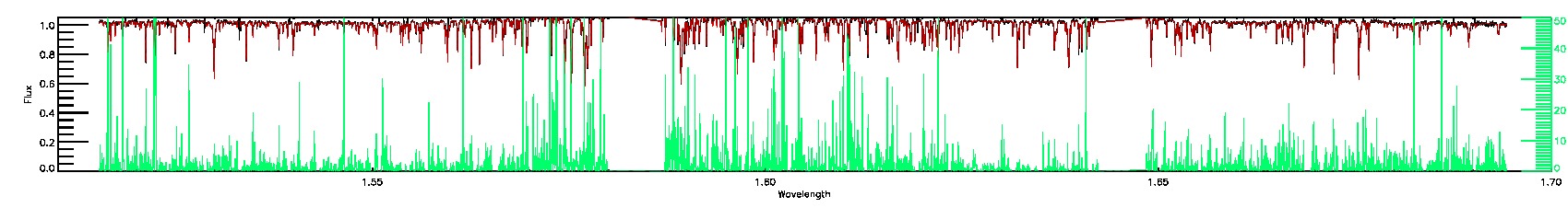
170-60
| 4847. | +/- | 6. |
| 4915. | +/- | 92. |
| 2.70 | +/- | 0. |
| 2.39 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.18 | +/- | 0. |
| 3.84 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.40 |
| -9999.00 |
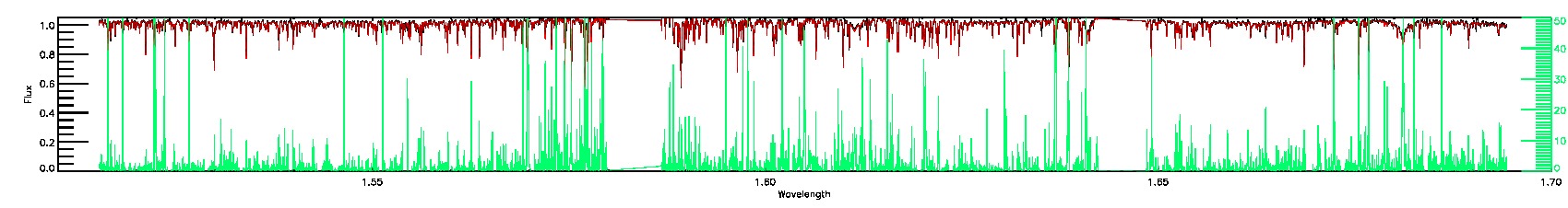
170-60
| 5955. | +/- | 12. |
| 5870. | +/- | 126. |
| 4.46 | +/- | 0. |
| 4.29 | +/- | 0. |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 5.19 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.44 |
| -9999.00 |

SUSPECT_BROAD_LINES
170-60
| 4035. | +/- | 5. |
| 4138. | +/- | 84. |
| 4.30 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 12.84 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
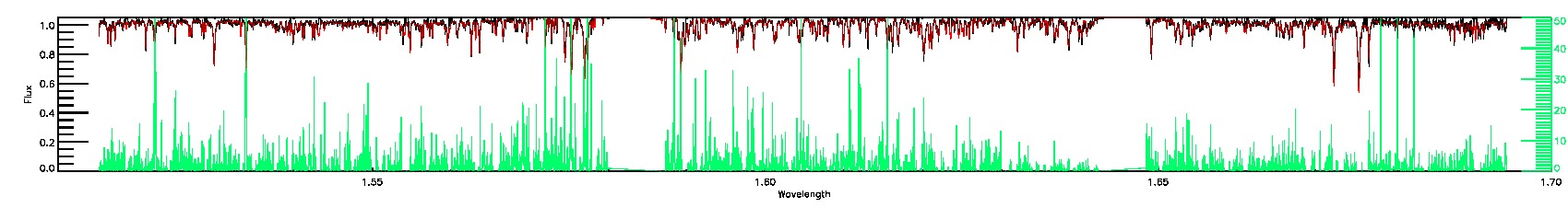
170-60
| 6031. | +/- | 14. |
| 5946. | +/- | 125. |
| 4.43 | +/- | 0. |
| 4.32 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.24 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
170-60
| 4992. | +/- | 9. |
| 5048. | +/- | 99. |
| 2.68 | +/- | 0. |
| 2.37 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -0.59 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.19 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.70 |
| -9999.00 |

PERSIST_LOW
STAR_WARN,COLORTE_WARN
170-60
| 3601. | +/- | 2. |
| 3690. | +/- | 64. |
| 4.51 | +/- | 0. |
| 4.83 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.97 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.47 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW
170-60
| 4800. | +/- | 6. |
| 4854. | +/- | 108. |
| 3.10 | +/- | 0. |
| 2.91 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.95 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.25 |
| -9999.00 |
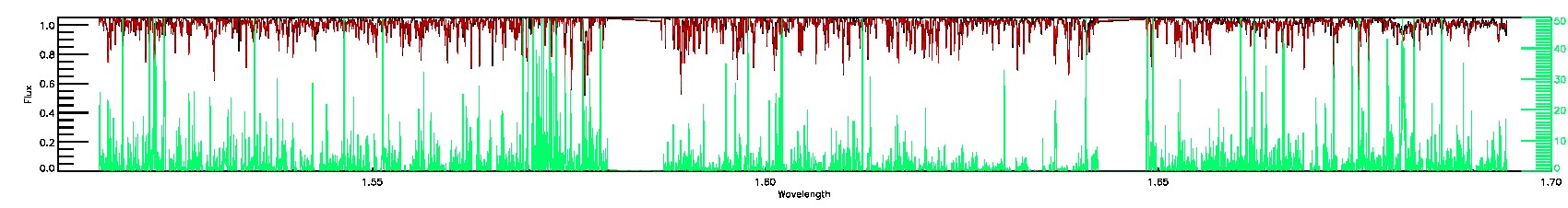
170-60
| 5428. | +/- | 18. |
| 5408. | +/- | 133. |
| 4.54 | +/- | 0. |
| 4.50 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.51 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.48 |
| -9999.00 |

170-60
| 4650. | +/- | 7. |
| 4721. | +/- | 99. |
| 4.52 | +/- | 0. |
| 4.64 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.41 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |

SUSPECT_BROAD_LINES
170-60
| 4161. | +/- | 3. |
| 4252. | +/- | 72. |
| 4.25 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.73 | +/- | 0. |
| 0.73 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 27.78 | +/- | 0. |
| 1.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |

170-60
| 5558. | +/- | 14. |
| 5531. | +/- | 110. |
| 4.25 | +/- | 0. |
| 4.27 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 4.74 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
170-60
| 3437. | +/- | 3. |
| 3526. | +/- | 71. |
| 4.61 | +/- | 0. |
| 4.96 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 6.48 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.01 |
| -9999.00 |
PERSIST_LOW
170-60
| 5951. | +/- | 12. |
| 5874. | +/- | 122. |
| 4.69 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 6.74 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.08 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_MED,PERSIST_LOW
170-60
| 5859. | +/- | 23. |
| 5807. | +/- | 157. |
| 4.35 | +/- | 0. |
| 4.41 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.91 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.87 |
| -9999.00 |

170-60
| 5861. | +/- | 11. |
| 5793. | +/- | 118. |
| 4.06 | +/- | 0. |
| 3.96 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.06 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |

PERSIST_LOW
170-60
| 6075. | +/- | 13. |
| 5978. | +/- | 125. |
| 4.06 | +/- | 0. |
| 3.88 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.68 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.00 | +/- | 0. |
| 5.72 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.04 |
| -9999.00 |
170-60
| 5308. | +/- | 14. |
| 5330. | +/- | 109. |
| 3.75 | +/- | 0. |
| 3.86 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -0.64 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.23 | +/- | 0. |
| 4.31 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.49 |
| -9999.00 |
170-60
| 5434. | +/- | 13. |
| 5418. | +/- | 116. |
| 4.64 | +/- | 0. |
| 4.64 | +/- | 0. |
| 1.79 | +/- | 0. |
| 1.79 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.66 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
170-60
| 5255. | +/- | 9. |
| 5252. | +/- | 109. |
| 4.56 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.18 |
| -9999.00 |
170-60
| 5915. | +/- | 25. |
| 5847. | +/- | 144. |
| 4.50 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.06 |
| -9999.00 |
PERSIST_LOW
170-60
| 6112. | +/- | 14. |
| 6012. | +/- | 127. |
| 4.49 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.87 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 4.04 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
170-60
| 5076. | +/- | 15. |
| 5105. | +/- | 129. |
| 4.73 | +/- | 0. |
| 4.84 | +/- | 0. |
| 1.90 | +/- | 0. |
| 1.90 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 11.40 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |

170-60
| 5017. | +/- | 12. |
| 5043. | +/- | 118. |
| 4.64 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 1.74 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.59 |
| -9999.00 |

170-60
| 5954. | +/- | 16. |
| 5872. | +/- | 137. |
| 4.22 | +/- | 0. |
| 4.08 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.61 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
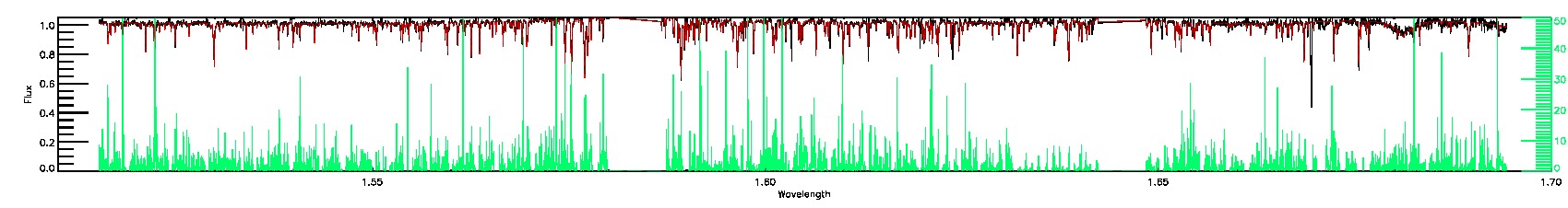
PERSIST_LOW
170-60
| 4096. | +/- | 4. |
| 4192. | +/- | 93. |
| 4.35 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 5.82 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.00 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
170-60
| 3528. | +/- | 5. |
| -10000. | +/- | -NaN |
| 5.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 29.51 | +/- | 0. |
| 1.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.29 |
| -9999.00 |
PERSIST_LOW
STAR_WARN,COLORTE_WARN,SN_WARN
170-60
| 3794. | +/- | 6. |
| 3900. | +/- | 90. |
| 4.49 | +/- | 0. |
| 4.93 | +/- | 0. |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 6.13 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.27 |
| -9999.00 |
170-60
| 5827. | +/- | 16. |
| 5778. | +/- | 122. |
| 4.46 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.19 | +/- | 0. |
| 1.60 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.07 |
| -9999.00 |

170-60
| 4635. | +/- | 4. |
| 4718. | +/- | 88. |
| 2.77 | +/- | 0. |
| 2.59 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.42 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.40 |
| -9999.00 |

170-60
| 4949. | +/- | 9. |
| 5027. | +/- | 101. |
| 2.98 | +/- | 0. |
| 2.91 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -0.98 | +/- | 0. |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.16 | +/- | 0. |
| 5.26 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.72 |
| -9999.00 |

PERSIST_LOW
170-60
| 3941. | +/- | 3. |
| 4051. | +/- | 91. |
| 4.39 | +/- | 0. |
| 4.84 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.86 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
170-60
| 5484. | +/- | 17. |
| 5461. | +/- | 133. |
| 4.04 | +/- | 0. |
| 4.03 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.09 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.43 |
| -9999.00 |

PERSIST_LOW
170-60
| 5792. | +/- | 16. |
| 5741. | +/- | 146. |
| 4.41 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.15 | +/- | 0. |
| 1.87 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 4752. | +/- | 5. |
| 4822. | +/- | 87. |
| 3.07 | +/- | 0. |
| 2.91 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
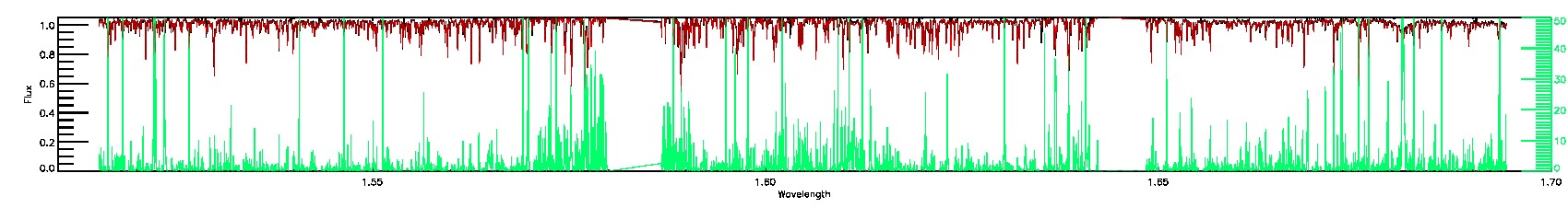
PERSIST_LOW
170-60
| 5952. | +/- | 12. |
| 5872. | +/- | 122. |
| 4.24 | +/- | 0. |
| 4.12 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 7.61 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.15 |
| -9999.00 |
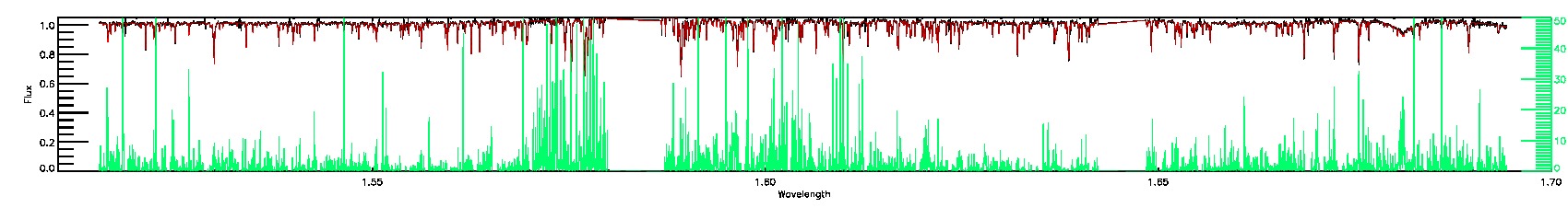
PERSIST_LOW
STAR_WARN,COLORTE_WARN
170-60
| 3706. | +/- | 2. |
| 3804. | +/- | 64. |
| 4.50 | +/- | 0. |
| 4.89 | +/- | 0. |
| 0.70 | +/- | 0. |
| 0.70 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.83 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
BRIGHT_NEIGHBOR
170-60
| 3928. | +/- | 2. |
| 4027. | +/- | 70. |
| 4.32 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 6.91 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
PERSIST_LOW
STAR_WARN,SN_WARN
170-60
| 5430. | +/- | 21. |
| 5408. | +/- | 144. |
| 4.57 | +/- | 0. |
| 4.51 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.31 |
| -9999.00 |

PERSIST_LOW
170-60
| 5768. | +/- | 20. |
| 5738. | +/- | 129. |
| 4.10 | +/- | 0. |
| 4.28 | +/- | 0. |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -0.65 | +/- | 0. |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.23 | +/- | 0. |
| 4.34 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.40 |
| -9999.00 |

PERSIST_MED,PERSIST_LOW,SUSPECT_BROAD_LINES
170-60
| 6308. | +/- | 19. |
| 6187. | +/- | 135. |
| 4.67 | +/- | 0. |
| 4.48 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 39.93 | +/- | 0. |
| 1.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
PERSIST_LOW
170-60
| 4565. | +/- | 3. |
| 4639. | +/- | 87. |
| 2.95 | +/- | 0. |
| 2.73 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.70 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.29 |
| -9999.00 |
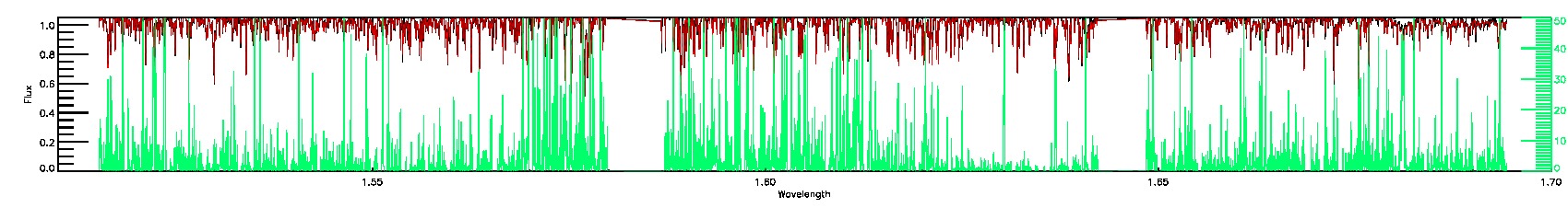
PERSIST_LOW
STAR_WARN,SN_WARN
170-60
| 5374. | +/- | 24. |
| 5381. | +/- | 143. |
| 4.43 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.61 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
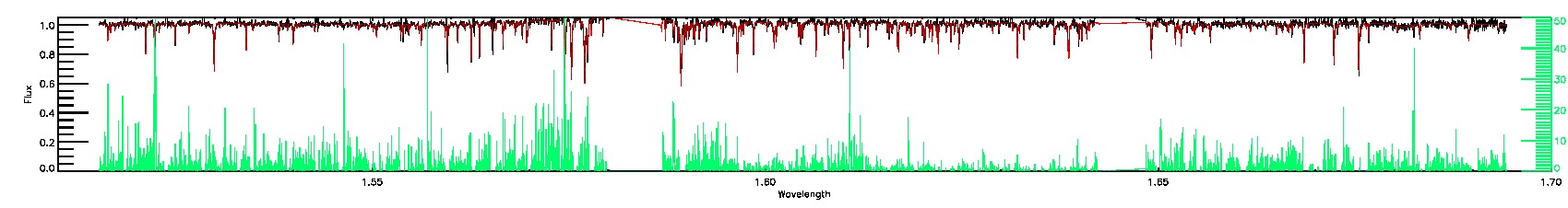
PERSIST_MED,PERSIST_LOW
STAR_WARN,SN_WARN
170-60
| 5468. | +/- | 18. |
| 5445. | +/- | 144. |
| 4.47 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.74 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.14 |
| -9999.00 |
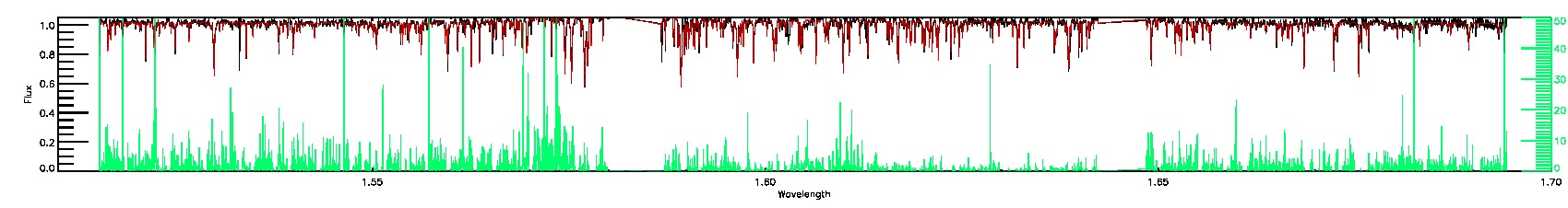
PERSIST_LOW
170-60
| 5030. | +/- | 9. |
| 5088. | +/- | 112. |
| 2.75 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -0.73 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.24 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.68 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
170-60
| 4548. | +/- | 9. |
| 4672. | +/- | 98. |
| 3.56 | +/- | 0. |
| 3.61 | +/- | 0. |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| -0.96 | +/- | 0. |
| -0.96 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.17 | +/- | 0. |
| 78.61 | +/- | 0. |
| 1.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.70 |
| -9999.00 |
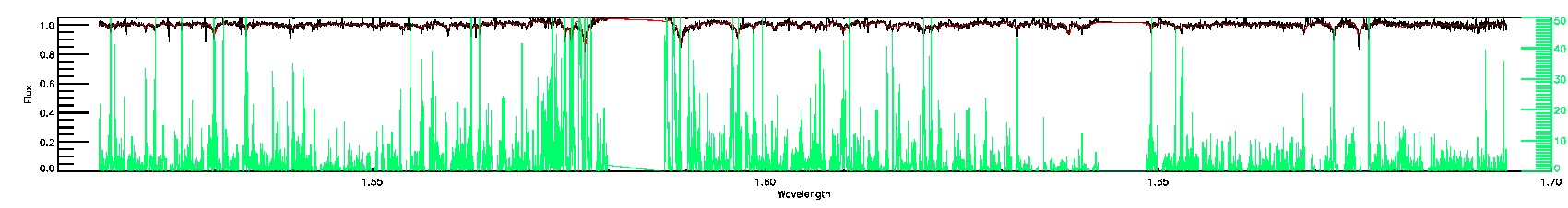
PERSIST_LOW
170-60
| 4282. | +/- | 3. |
| 4388. | +/- | 90. |
| 4.39 | +/- | 0. |
| 4.74 | +/- | 0. |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.41 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.79 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.29 |
| -9999.00 |
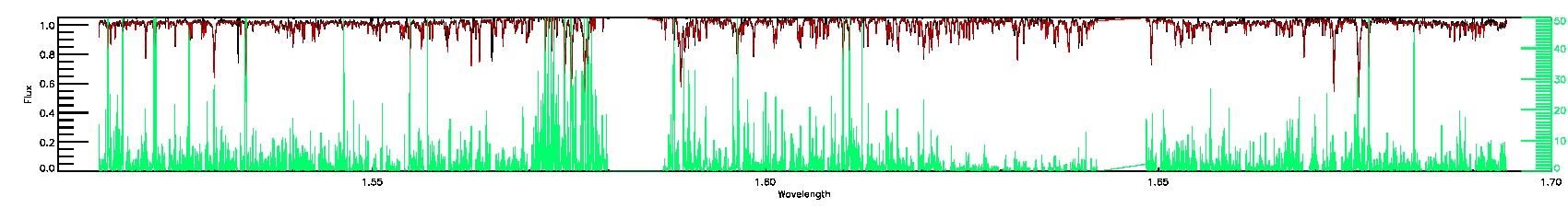
PERSIST_LOW
170-60
| 5306. | +/- | 11. |
| 5299. | +/- | 125. |
| 4.57 | +/- | 0. |
| 4.56 | +/- | 0. |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.90 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.06 |
| -9999.00 |
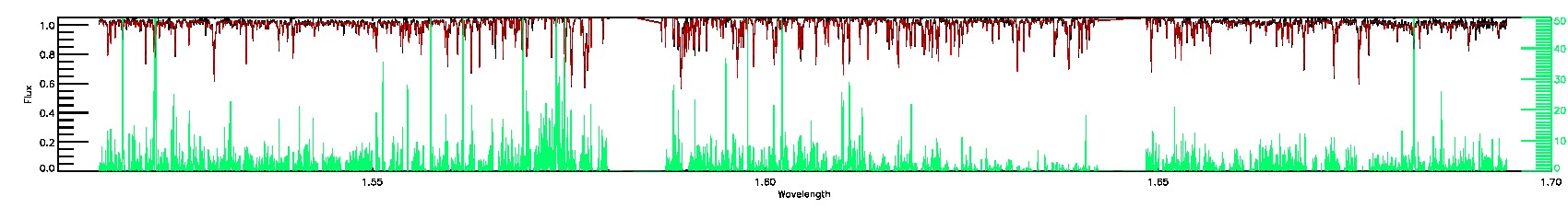
BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 5687. | +/- | 12. |
| 5635. | +/- | 111. |
| 4.33 | +/- | 0. |
| 4.24 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
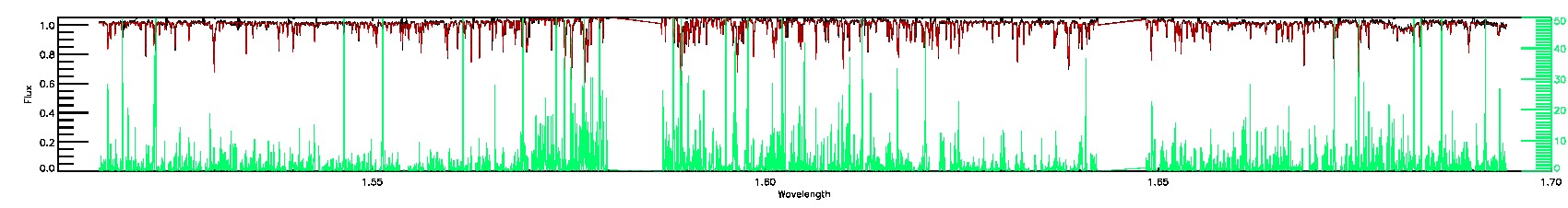
BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 4048. | +/- | 5. |
| 4152. | +/- | 94. |
| 4.36 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.37 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.24 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.36 |
| -9999.00 |
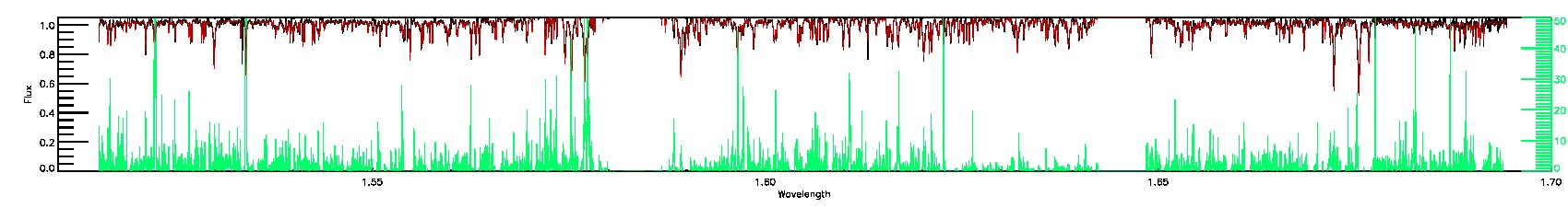
PERSIST_LOW
170-60
| 5188. | +/- | 8. |
| 5197. | +/- | 97. |
| 3.90 | +/- | 0. |
| 3.90 | +/- | 0. |
| 0.95 | +/- | 0. |
| 0.95 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.99 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.02 |
| -9999.00 |
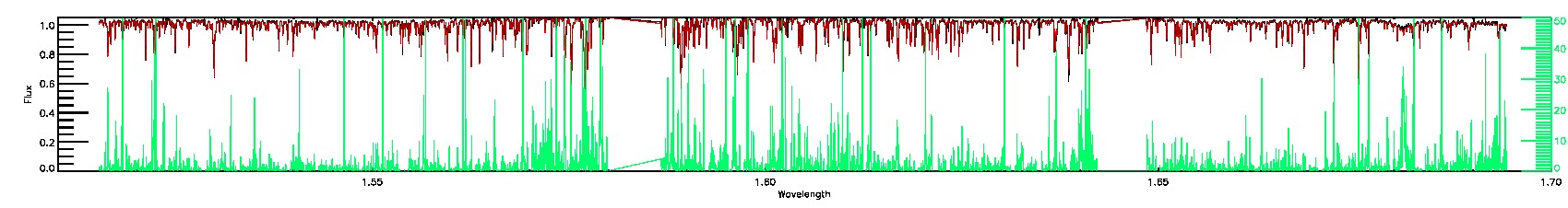
PERSIST_LOW
170-60
| 4499. | +/- | 3. |
| 4574. | +/- | 79. |
| 4.47 | +/- | 0. |
| 4.49 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.09 | +/- | 0. |
| 1.52 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.33 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
STAR_WARN,SN_WARN
170-60
| 5078. | +/- | 23. |
| 5138. | +/- | 140. |
| 3.20 | +/- | 0. |
| 2.83 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.89 | +/- | 0. |
| -0.89 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.20 | +/- | 0. |
| 4.99 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.69 |
| -9999.00 |
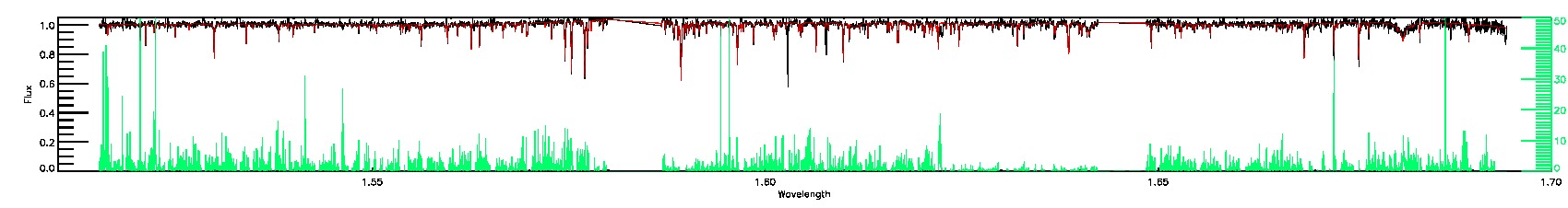
170-60
| 4929. | +/- | 11. |
| 4968. | +/- | 113. |
| 4.58 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.77 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
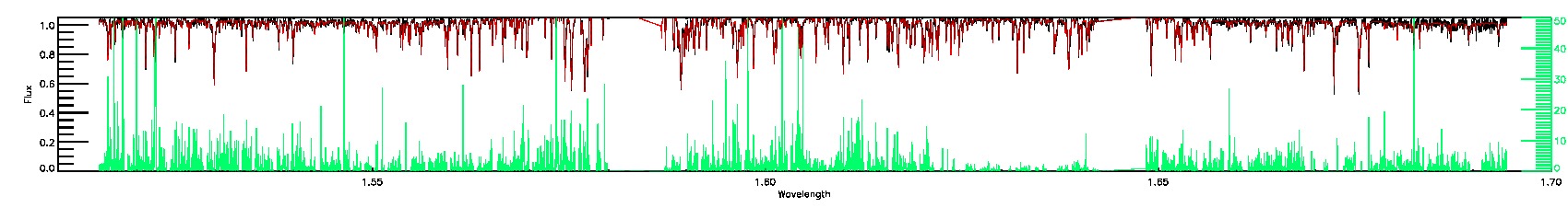
170-60
| 4478. | +/- | 6. |
| 4558. | +/- | 96. |
| 4.45 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.16 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |

170-60
| 4303. | +/- | 2. |
| 4395. | +/- | 75. |
| 4.32 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 5.56 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |

170-60
| 3768. | +/- | 2. |
| 3856. | +/- | 68. |
| 4.49 | +/- | 0. |
| 4.77 | +/- | 0. |
| 0.60 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.17 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
PERSIST_MED,PERSIST_LOW
170-60
| 5194. | +/- | 10. |
| 5207. | +/- | 120. |
| 4.60 | +/- | 0. |
| 4.67 | +/- | 0. |
| 2.08 | +/- | 0. |
| 2.08 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.99 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |

PERSIST_MED
170-60
| 5560. | +/- | 15. |
| 5528. | +/- | 133. |
| 4.13 | +/- | 0. |
| 4.09 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 2.28 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |

PERSIST_MED
170-60
| 5235. | +/- | 12. |
| 5258. | +/- | 105. |
| 4.34 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.54 | +/- | 0. |
| 0.54 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.00 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.30 |
| -9999.00 |

PERSIST_LOW
170-60
| 4805. | +/- | 5. |
| 4863. | +/- | 87. |
| 4.56 | +/- | 0. |
| 4.70 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
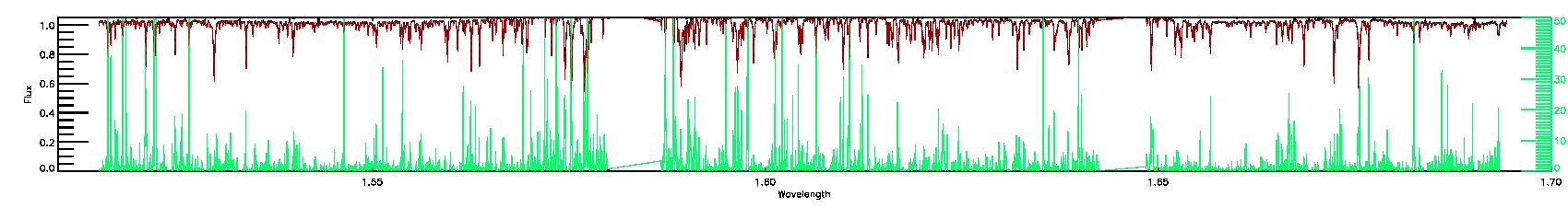
170-60
| 4707. | +/- | 5. |
| 4782. | +/- | 86. |
| 4.48 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.69 | +/- | 0. |
| 0.69 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.48 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.74 |
| -9999.00 |
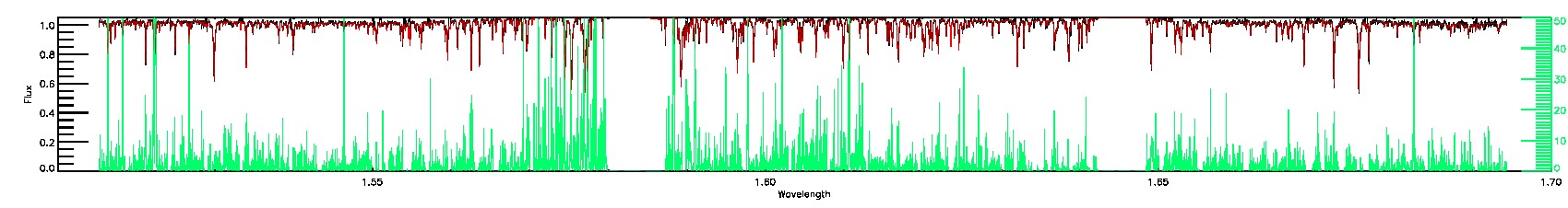
170-60
| 6173. | +/- | 35. |
| 6084. | +/- | 164. |
| 4.29 | +/- | 0. |
| 4.28 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 12.69 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
PERSIST_LOW
170-60
| 5946. | +/- | 23. |
| 5878. | +/- | 162. |
| 4.09 | +/- | 0. |
| 4.07 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.27 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.47 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |

170-60
| 5635. | +/- | 12. |
| 5591. | +/- | 110. |
| 4.12 | +/- | 0. |
| 4.05 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.06 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
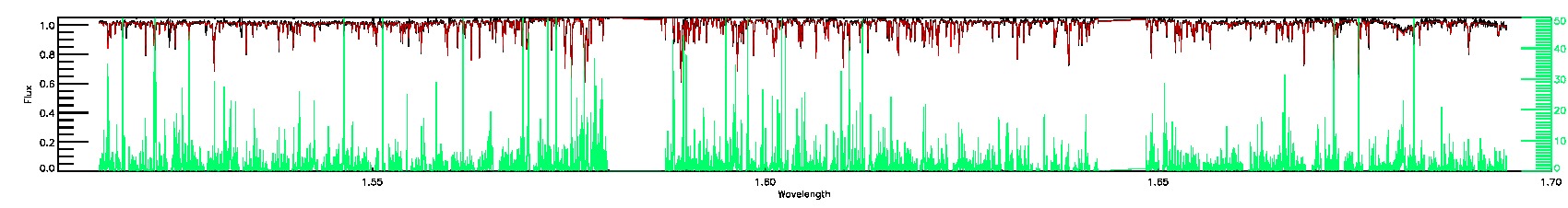
170-60
| 4882. | +/- | 6. |
| 4950. | +/- | 94. |
| 2.65 | +/- | 0. |
| 2.36 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.83 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -0.55 | +/- | 0. |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.14 | +/- | 0. |
| 4.09 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.04 |
| -9999.00 |
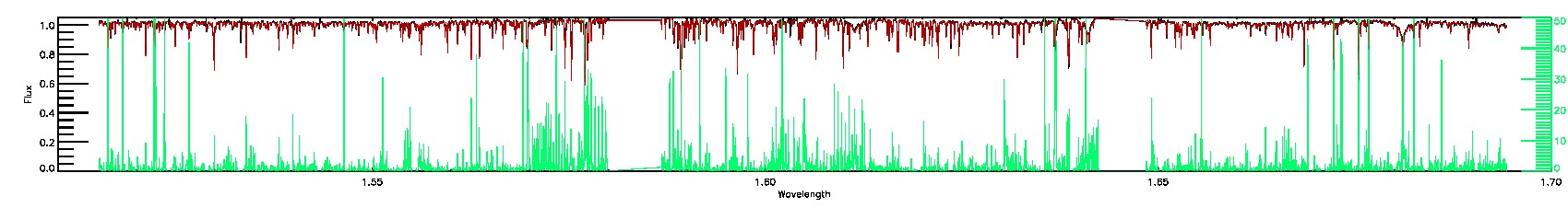
170-60
| 5181. | +/- | 8. |
| 5188. | +/- | 103. |
| 4.47 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.87 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.66 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.21 |
| -9999.00 |

PERSIST_LOW
170-60
| 4967. | +/- | 8. |
| 5016. | +/- | 94. |
| 4.55 | +/- | 0. |
| 4.74 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |

170-60
| 6404. | +/- | 14. |
| 6261. | +/- | 134. |
| 4.38 | +/- | 0. |
| 4.06 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 8.66 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.27 |
| -9999.00 |
170-60
| 4747. | +/- | 5. |
| 4807. | +/- | 94. |
| 3.16 | +/- | 0. |
| 2.97 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.95 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.24 |
| -9999.00 |

PERSIST_MED
170-60
| 4766. | +/- | 4. |
| 4819. | +/- | 84. |
| 3.24 | +/- | 0. |
| 3.05 | +/- | 0. |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.79 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.33 |
| -9999.00 |
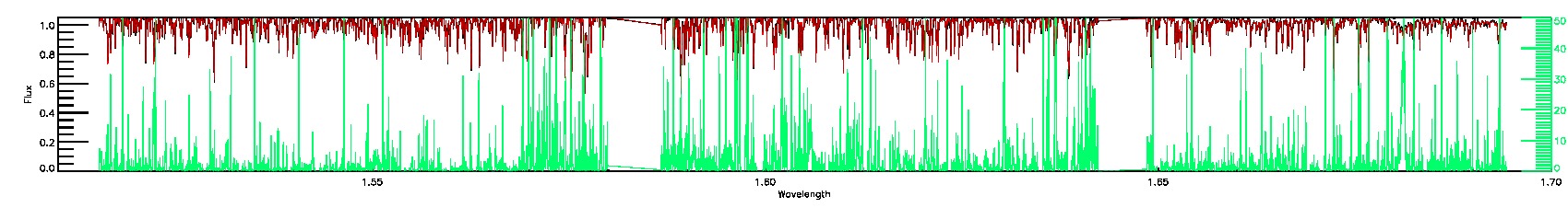
170-60
| 4716. | +/- | 7. |
| 4807. | +/- | 101. |
| 2.69 | +/- | 0. |
| 2.55 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -0.65 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.22 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.86 |
| -9999.00 |

170-60
| 5821. | +/- | 24. |
| 5769. | +/- | 143. |
| 4.26 | +/- | 0. |
| 4.28 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.56 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.13 |
| -9999.00 |

170-60
| 5654. | +/- | 15. |
| 5622. | +/- | 115. |
| 4.41 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.48 | +/- | 0. |
| 0.48 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.11 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.14 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5043. | +/- | 8. |
| 5074. | +/- | 104. |
| 3.58 | +/- | 0. |
| 3.48 | +/- | 0. |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.22 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.18 |
| -9999.00 |

STAR_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
170-60
| 3000. | +/- | 0. |
| -10000. | +/- | -NaN |
| 3.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.87 | +/- | 0. |
| 1.87 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.40 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.01 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 6063. | +/- | 23. |
| 5977. | +/- | 164. |
| 4.21 | +/- | 0. |
| 4.13 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.59 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
170-60
| 5241. | +/- | 11. |
| 5258. | +/- | 102. |
| 3.64 | +/- | 0. |
| 3.62 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.64 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.37 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 5399. | +/- | 13. |
| 5405. | +/- | 131. |
| 4.53 | +/- | 0. |
| 4.72 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -0.52 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.19 | +/- | 0. |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.52 |
| -9999.00 |

PERSIST_MED
170-60
| 5540. | +/- | 8. |
| 5499. | +/- | 114. |
| 4.63 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.65 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.36 |
| -9999.00 |
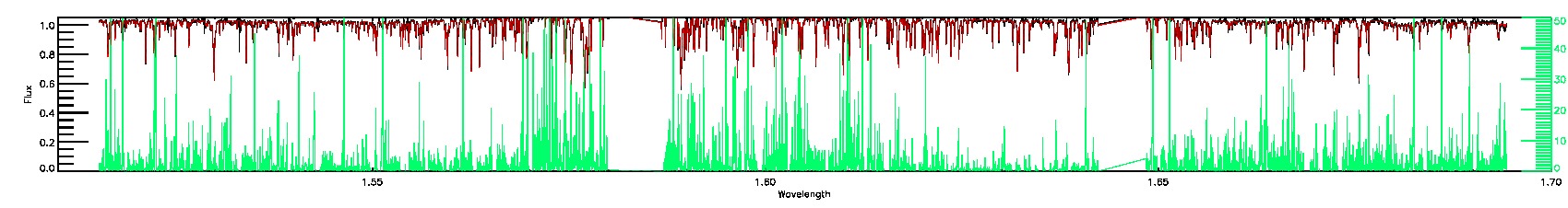
170-60
| 5597. | +/- | 11. |
| 5566. | +/- | 111. |
| 4.54 | +/- | 0. |
| 4.55 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.59 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
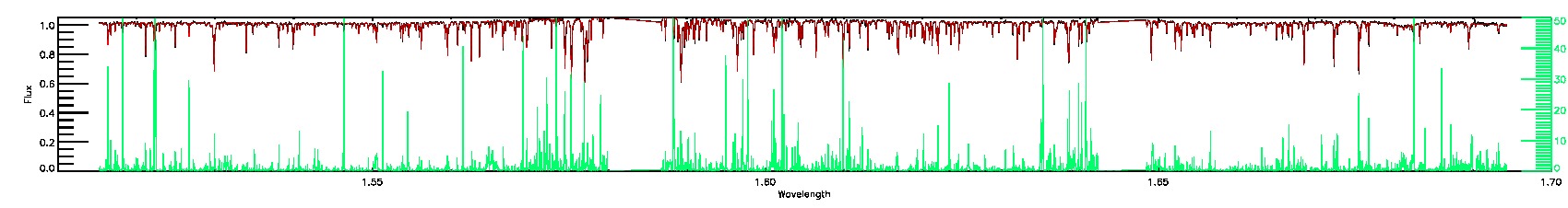
PERSIST_LOW
170-60
| 4769. | +/- | 4. |
| 4822. | +/- | 84. |
| 3.28 | +/- | 0. |
| 3.10 | +/- | 0. |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.78 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.22 |
| -9999.00 |

170-60
| 5931. | +/- | 27. |
| 5874. | +/- | 141. |
| 3.85 | +/- | 0. |
| 3.87 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 6.50 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.06 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5794. | +/- | 17. |
| 5743. | +/- | 131. |
| 4.42 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.75 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.47 |
| -9999.00 |
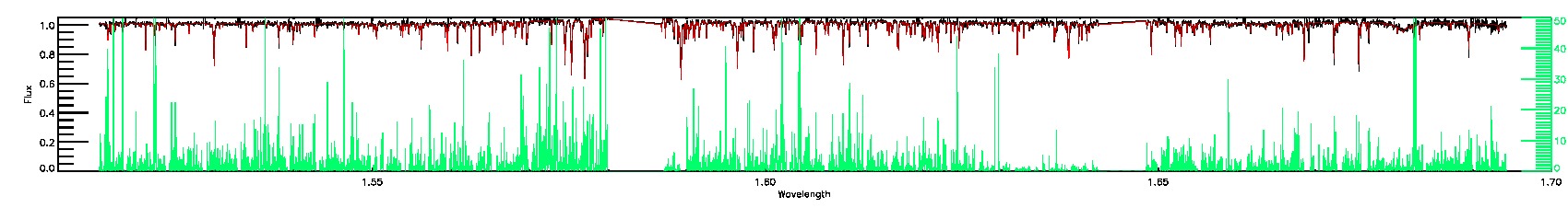
170-60
| 5962. | +/- | 13. |
| 5886. | +/- | 123. |
| 4.53 | +/- | 0. |
| 4.45 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.20 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.53 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.16 |
| -9999.00 |
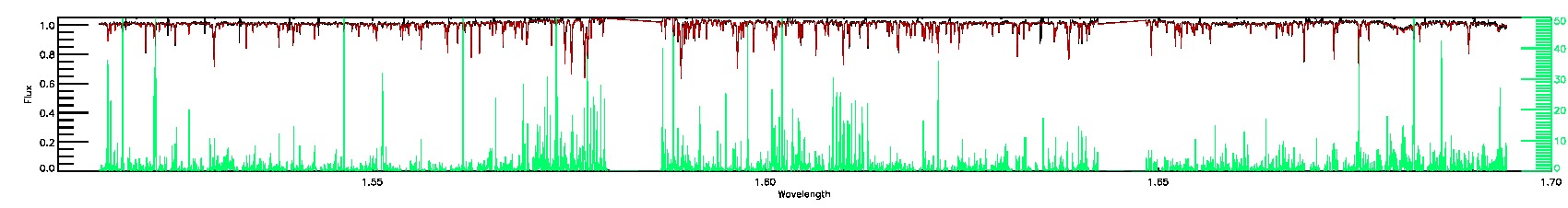
170-60
| 5811. | +/- | 14. |
| 5750. | +/- | 127. |
| 4.38 | +/- | 0. |
| 4.30 | +/- | 0. |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 2.66 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.29 |
| -9999.00 |
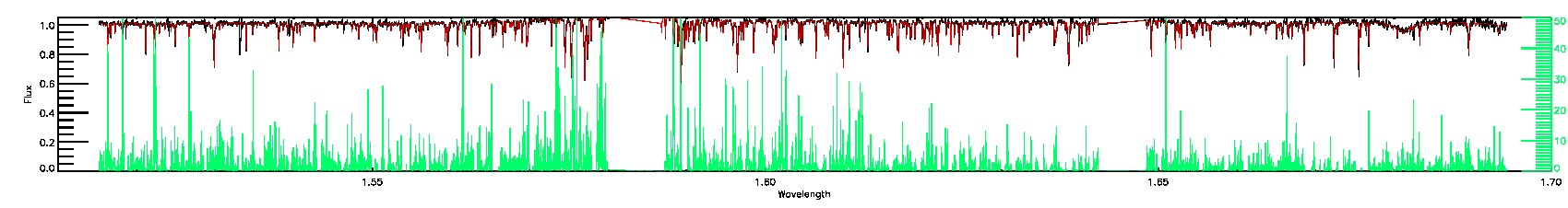
170-60
| 5112. | +/- | 10. |
| 5163. | +/- | 103. |
| 3.08 | +/- | 0. |
| 2.99 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -0.79 | +/- | 0. |
| -0.79 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.21 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.50 |
| -9999.00 |

SUSPECT_BROAD_LINES
170-60
| 6220. | +/- | 16. |
| 6107. | +/- | 130. |
| 4.38 | +/- | 0. |
| 4.19 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 16.51 | +/- | 0. |
| 1.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
170-60
| 4236. | +/- | 3. |
| 4328. | +/- | 79. |
| 4.39 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.53 | +/- | 0. |
| 0.53 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.25 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |

PERSIST_LOW
170-60
| 5469. | +/- | 11. |
| 5442. | +/- | 122. |
| 3.98 | +/- | 0. |
| 3.92 | +/- | 0. |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.86 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.00 |
| -9999.00 |
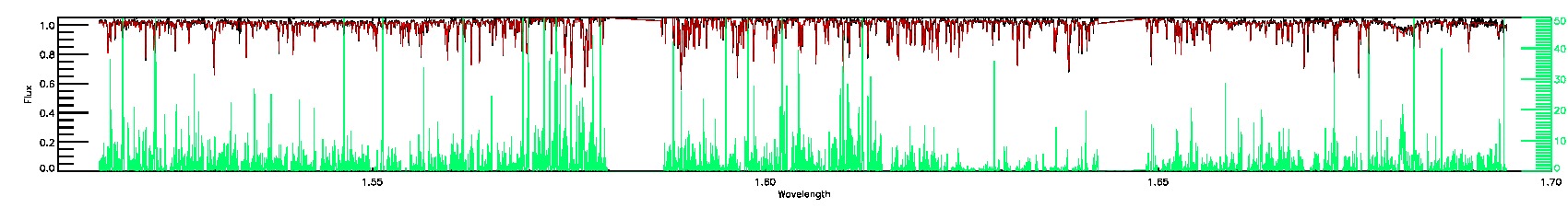
170-60
| 4844. | +/- | 6. |
| 4896. | +/- | 88. |
| 3.65 | +/- | 0. |
| 3.59 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.13 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |

SUSPECT_RV_COMBINATION
170-60
| 8501. | +/- | 29. |
| 8375. | +/- | 317. |
| 4.43 | +/- | 0. |
| 4.74 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 34.25 | +/- | 0. |
| 1.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.02 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.94 |
| -9999.00 |
PERSIST_LOW
170-60
| 4580. | +/- | 3. |
| 4657. | +/- | 80. |
| 2.88 | +/- | 0. |
| 2.66 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.89 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.16 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
170-60
| 4648. | +/- | 12. |
| 4748. | +/- | 113. |
| 3.59 | +/- | 0. |
| 3.62 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -0.69 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 8.20 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.36 |
| -9999.00 |

PERSIST_MED
170-60
| 4670. | +/- | 5. |
| 4758. | +/- | 87. |
| 2.67 | +/- | 0. |
| 2.51 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.87 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.51 |
| -9999.00 |
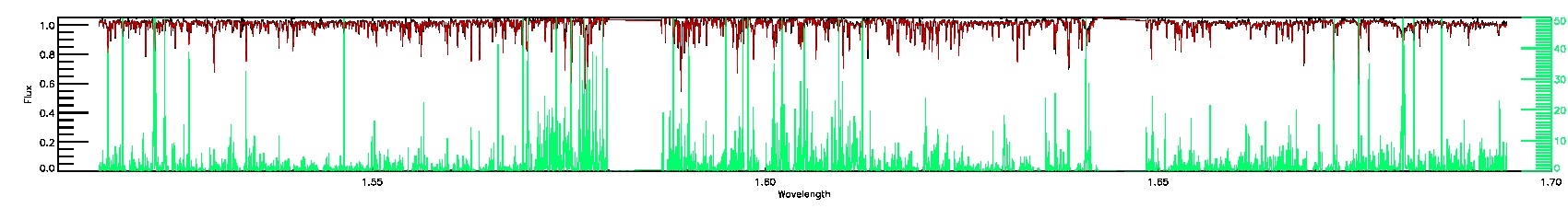
170-60
| 5264. | +/- | 9. |
| 5268. | +/- | 106. |
| 4.66 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.69 | +/- | 0. |
| 0.69 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.76 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5752. | +/- | 35. |
| 5714. | +/- | 148. |
| 3.84 | +/- | 0. |
| 3.87 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.81 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.75 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5337. | +/- | 11. |
| 5326. | +/- | 115. |
| 3.89 | +/- | 0. |
| 3.83 | +/- | 0. |
| 0.66 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.33 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.17 |
| -9999.00 |

170-60
| 6151. | +/- | 14. |
| 6042. | +/- | 127. |
| 4.30 | +/- | 0. |
| 4.09 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.78 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.09 |
| -9999.00 |
170-60
| 4710. | +/- | 6. |
| 4763. | +/- | 95. |
| 4.52 | +/- | 0. |
| 4.53 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.79 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.33 |
| -9999.00 |

170-60
| 5847. | +/- | 13. |
| 5780. | +/- | 125. |
| 4.38 | +/- | 0. |
| 4.28 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.04 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.21 |
| -9999.00 |

170-60
| 4944. | +/- | 6. |
| 4981. | +/- | 90. |
| 3.76 | +/- | 0. |
| 3.73 | +/- | 0. |
| 0.56 | +/- | 0. |
| 0.56 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 2.98 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.18 |
| -9999.00 |
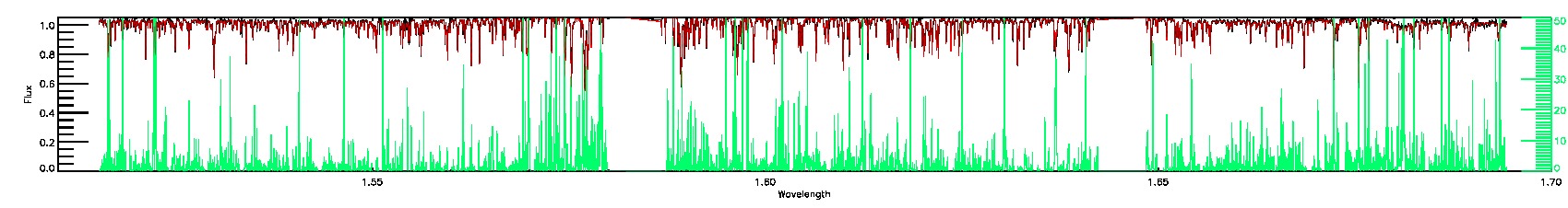
170-60
| 4497. | +/- | 7. |
| 4612. | +/- | 106. |
| 3.90 | +/- | 0. |
| 4.21 | +/- | 0. |
| 0.74 | +/- | 0. |
| 0.74 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -0.63 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.07 | +/- | 0. |
| 6.16 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.30 |
| -9999.00 |

170-60
| 4821. | +/- | 5. |
| 4870. | +/- | 86. |
| 2.94 | +/- | 0. |
| 2.57 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.88 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.21 |
| -9999.00 |

170-60
| 5452. | +/- | 10. |
| 5431. | +/- | 105. |
| 4.56 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.98 | +/- | 0. |
| 0.98 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.33 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |

PERSIST_LOW
170-60
| 5366. | +/- | 22. |
| 5358. | +/- | 135. |
| 4.57 | +/- | 0. |
| 4.59 | +/- | 0. |
| 1.79 | +/- | 0. |
| 1.79 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.09 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.98 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.25 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 4950. | +/- | 9. |
| 4997. | +/- | 104. |
| 4.32 | +/- | 0. |
| 4.48 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.07 | +/- | 0. |
| 2.50 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.41 |
| -9999.00 |

170-60
| 5862. | +/- | 12. |
| 5798. | +/- | 120. |
| 4.50 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.57 | +/- | 0. |
| 0.57 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
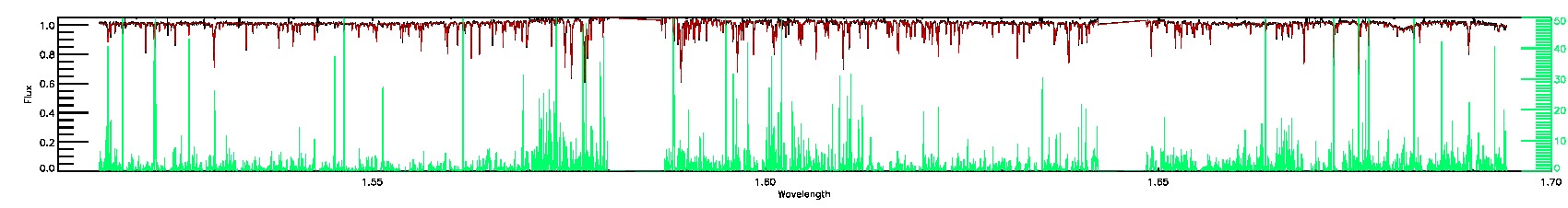
170-60
| 4790. | +/- | 4. |
| 4835. | +/- | 84. |
| 3.52 | +/- | 0. |
| 3.34 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.22 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.61 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.35 |
| -9999.00 |

170-60
| 5808. | +/- | 18. |
| 5746. | +/- | 137. |
| 4.28 | +/- | 0. |
| 4.19 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 8.76 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.06 |
| -9999.00 |

170-60
| 6473. | +/- | 17. |
| 6338. | +/- | 144. |
| 4.43 | +/- | 0. |
| 4.26 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 12.67 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.65 |
| -9999.00 |
170-60
| 6074. | +/- | 14. |
| 5983. | +/- | 127. |
| 4.49 | +/- | 0. |
| 4.38 | +/- | 0. |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.34 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
170-60
| 4060. | +/- | 4. |
| 4147. | +/- | 83. |
| 4.36 | +/- | 0. |
| 4.57 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.36 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.20 |
| -9999.00 |

170-60
| 5458. | +/- | 14. |
| 5451. | +/- | 109. |
| 4.17 | +/- | 0. |
| 4.28 | +/- | 0. |
| 0.48 | +/- | 0. |
| 0.48 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.29 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.35 |
| -9999.00 |

PERSIST_LOW
170-60
| 6225. | +/- | 15. |
| 6113. | +/- | 131. |
| 4.68 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.34 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
170-60
| 6047. | +/- | 18. |
| 5962. | +/- | 144. |
| 4.29 | +/- | 0. |
| 4.21 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.94 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.51 |
| -9999.00 |
BRIGHT_NEIGHBOR
170-60
| 5689. | +/- | 10. |
| 5632. | +/- | 110. |
| 4.39 | +/- | 0. |
| 4.24 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.77 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.35 |
| -9999.00 |
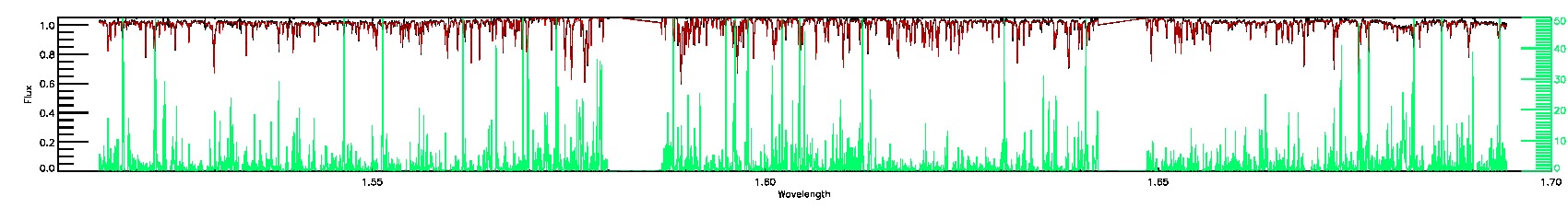
BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 5028. | +/- | 15. |
| 5079. | +/- | 117. |
| 3.16 | +/- | 0. |
| 3.06 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -0.56 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.11 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.35 |
| -9999.00 |

170-60
| 6117. | +/- | 14. |
| 6014. | +/- | 126. |
| 4.37 | +/- | 0. |
| 4.18 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 6.90 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.20 |
| -9999.00 |
170-60
| 4869. | +/- | 6. |
| 4934. | +/- | 93. |
| 2.71 | +/- | 0. |
| 2.39 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.90 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.50 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 4173. | +/- | 3. |
| 4270. | +/- | 77. |
| 4.38 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.66 | +/- | 0. |
| 0.66 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 3.11 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
170-60
| 5771. | +/- | 22. |
| 5736. | +/- | 123. |
| 4.02 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -0.57 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.41 | +/- | 0. |
| 17.40 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.41 |
| -9999.00 |

170-60
| 4625. | +/- | 5. |
| 4717. | +/- | 87. |
| 4.42 | +/- | 0. |
| 4.71 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.69 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.41 |
| -9999.00 |

170-60
| 6081. | +/- | 16. |
| 5992. | +/- | 128. |
| 4.27 | +/- | 0. |
| 4.17 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.27 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.76 |
| -9999.00 |
170-60
| 5072. | +/- | 9. |
| 5089. | +/- | 109. |
| 4.21 | +/- | 0. |
| 4.20 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.01 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.12 |
| -9999.00 |

170-60
| 5902. | +/- | 10. |
| 5825. | +/- | 118. |
| 4.49 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.92 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
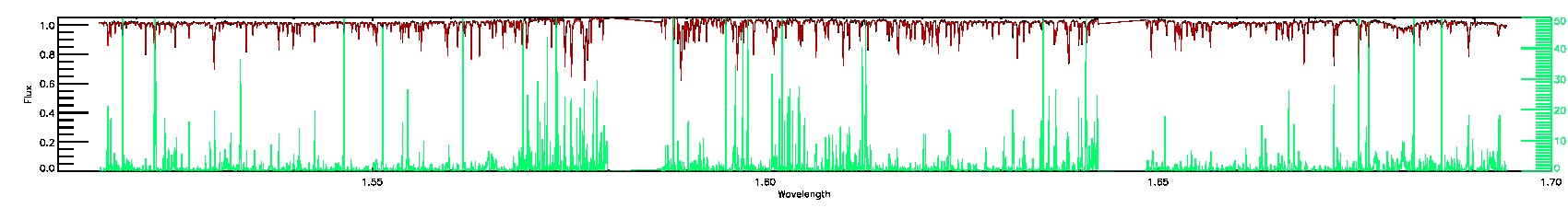
170-60
| 5217. | +/- | 23. |
| 5244. | +/- | 127. |
| 3.42 | +/- | 0. |
| 3.12 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -0.52 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.02 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.70 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5950. | +/- | 15. |
| 5873. | +/- | 132. |
| 4.23 | +/- | 0. |
| 4.13 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.06 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.31 |
| -9999.00 |
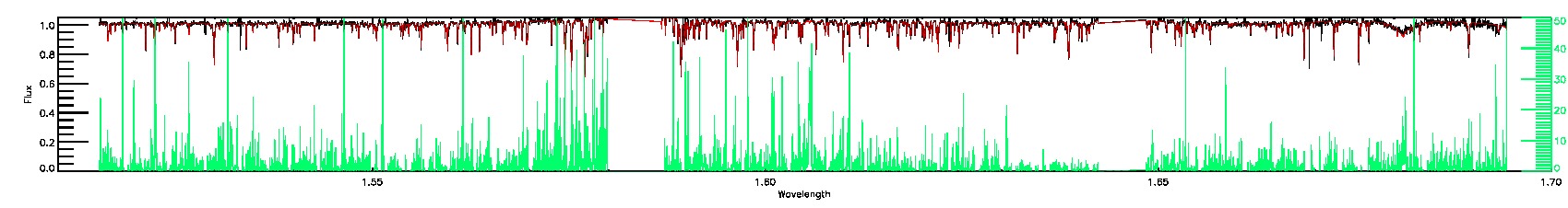
PERSIST_LOW,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
170-60
| 4976. | +/- | 17. |
| 5042. | +/- | 99. |
| 3.95 | +/- | 0. |
| 4.28 | +/- | 0. |
| 4.35 | +/- | 0. |
| 4.35 | +/- | -NaN |
| -0.77 | +/- | 0. |
| -0.77 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 52.34 | +/- | 0. |
| 1.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.77 |
| -9999.00 |

170-60
| 5054. | +/- | 26. |
| 5121. | +/- | 131. |
| 3.25 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| -1.00 | +/- | 0. |
| -1.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.30 | +/- | 0. |
| 5.31 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.34 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5967. | +/- | 19. |
| 5906. | +/- | 129. |
| 4.30 | +/- | 0. |
| 4.37 | +/- | 0. |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.15 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.56 |
| -9999.00 |

170-60
| 5580. | +/- | 14. |
| 5563. | +/- | 114. |
| 3.81 | +/- | 0. |
| 3.86 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.90 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.36 |
| -9999.00 |
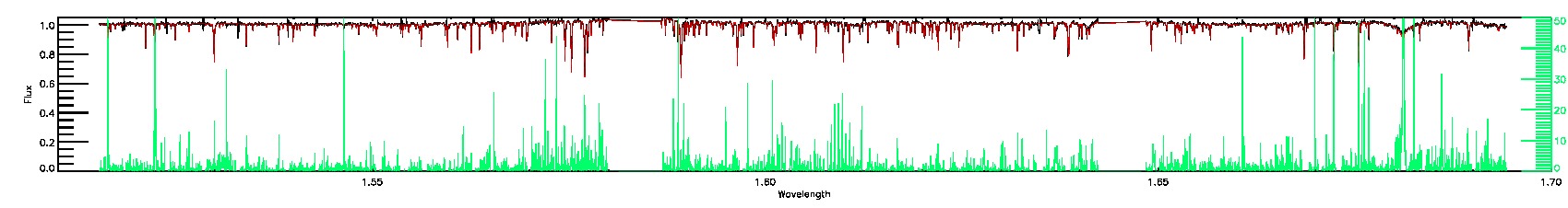
SUSPECT_BROAD_LINES
170-60
| 5842. | +/- | 13. |
| 5782. | +/- | 119. |
| 4.46 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.98 | +/- | 0. |
| 0.98 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 15.69 | +/- | 0. |
| 1.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
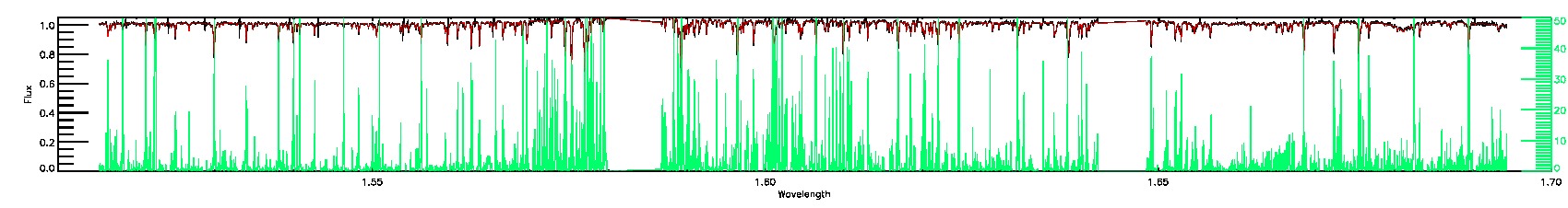
BRIGHT_NEIGHBOR
170-60
| 5936. | +/- | 12. |
| 5862. | +/- | 122. |
| 4.30 | +/- | 0. |
| 4.23 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.78 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
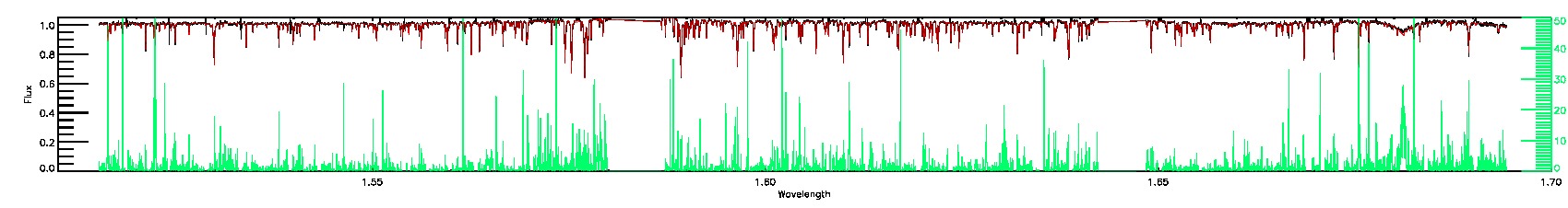
170-60
| 6288. | +/- | 17. |
| 6175. | +/- | 136. |
| 4.46 | +/- | 0. |
| 4.34 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 7.50 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
PERSIST_LOW
170-60
| 3829. | +/- | 3. |
| 3923. | +/- | 78. |
| 4.45 | +/- | 0. |
| 4.77 | +/- | 0. |
| 0.47 | +/- | 0. |
| 0.47 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.58 |
| -9999.00 |
170-60
| 5651. | +/- | 15. |
| 5605. | +/- | 124. |
| 4.44 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.97 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -2.37 |
| -9999.00 |
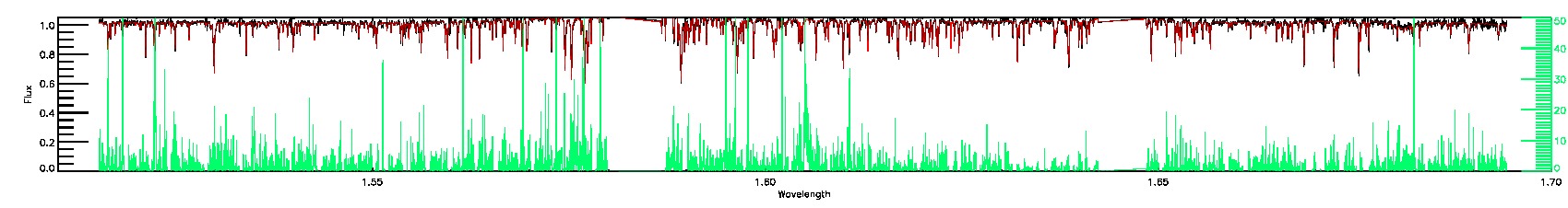
BRIGHT_NEIGHBOR
170-60
| 5934. | +/- | 20. |
| 5868. | +/- | 136. |
| 4.32 | +/- | 0. |
| 4.32 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.29 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
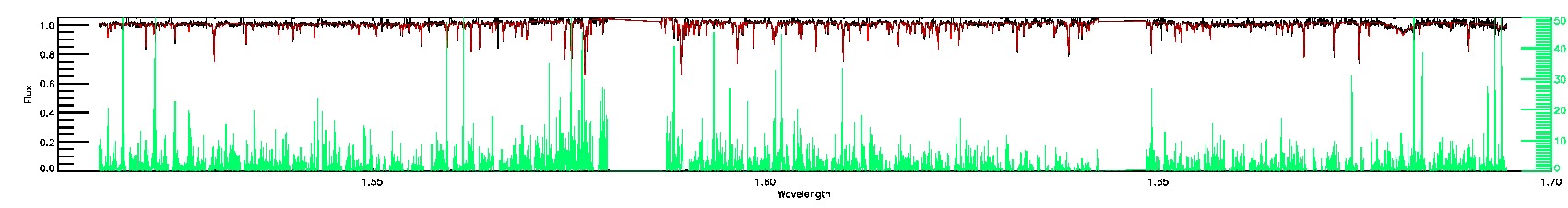
170-60
| 6202. | +/- | 17. |
| 6092. | +/- | 130. |
| 4.43 | +/- | 0. |
| 4.26 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 8.34 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.75 |
| -9999.00 |
170-60
| 5197. | +/- | 15. |
| 5220. | +/- | 117. |
| 3.57 | +/- | 0. |
| 3.51 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.66 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.46 |
| -9999.00 |

PERSIST_LOW
170-60
| 5584. | +/- | 11. |
| 5552. | +/- | 110. |
| 4.35 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.10 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
170-60
| 5603. | +/- | 10. |
| 5559. | +/- | 108. |
| 4.26 | +/- | 0. |
| 4.15 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.14 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.26 |
| -9999.00 |
PERSIST_LOW
170-60
| 4938. | +/- | 11. |
| 4981. | +/- | 120. |
| 4.41 | +/- | 0. |
| 4.53 | +/- | 0. |
| 0.58 | +/- | 0. |
| 0.58 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.12 | +/- | 0. |
| 1.73 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.16 |
| -9999.00 |

STAR_BAD
170-60
| 3973. | +/- | 2. |
| -10000. | +/- | -NaN |
| 4.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.30 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.17 |
| -9999.00 |
PERSIST_LOW
170-60
| 5462. | +/- | 18. |
| 5451. | +/- | 133. |
| 4.48 | +/- | 0. |
| 4.56 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5590. | +/- | 10. |
| 5546. | +/- | 107. |
| 4.75 | +/- | 0. |
| 4.63 | +/- | 0. |
| 0.86 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.25 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
170-60
| 3492. | +/- | 3. |
| 3596. | +/- | 70. |
| 4.23 | +/- | 0. |
| 4.71 | +/- | 0. |
| 1.12 | +/- | 0. |
| 1.12 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 6.24 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
SUSPECT_BROAD_LINES
170-60
| 5602. | +/- | 45. |
| 5575. | +/- | 142. |
| 4.25 | +/- | 0. |
| 4.31 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.53 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.13 | +/- | 0. |
| 49.20 | +/- | 0. |
| 1.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.44 |
| -9999.00 |

170-60
| 5915. | +/- | 24. |
| 5851. | +/- | 144. |
| 4.14 | +/- | 0. |
| 4.13 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.82 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |

PERSIST_LOW
170-60
| 4270. | +/- | 6. |
| 4361. | +/- | 97. |
| 4.40 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.72 | +/- | 0. |
| 0.72 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.22 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
170-60
| 4305. | +/- | 2. |
| 4387. | +/- | 73. |
| 2.50 | +/- | 0. |
| 2.25 | +/- | 0. |
| 1.12 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.16 |
| -9999.00 |

170-60
| 4517. | +/- | 3. |
| 4621. | +/- | 83. |
| 2.44 | +/- | 0. |
| 2.26 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -0.41 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.76 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.92 |
| -9999.00 |

170-60
| 5844. | +/- | 21. |
| 5781. | +/- | 143. |
| 4.34 | +/- | 0. |
| 4.29 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.81 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.06 |
| -9999.00 |
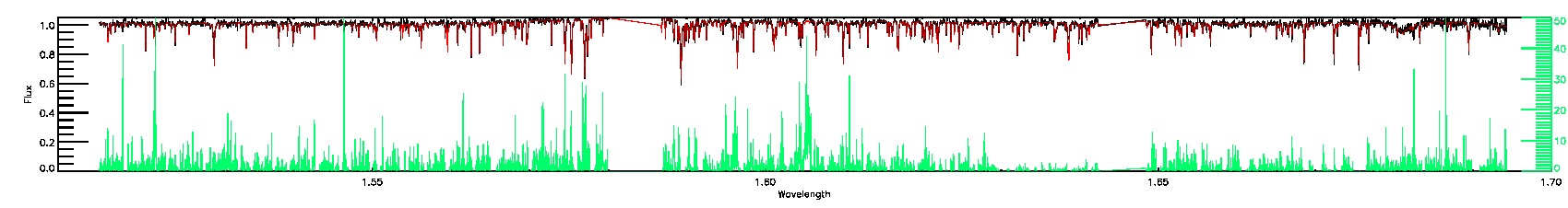
PERSIST_LOW
170-60
| 5835. | +/- | 19. |
| 5775. | +/- | 151. |
| 4.66 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.68 | +/- | 0. |
| 0.68 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.70 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |

170-60
| 5504. | +/- | 26. |
| 5490. | +/- | 138. |
| 4.42 | +/- | 0. |
| 4.51 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 1.68 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |

170-60
| 5676. | +/- | 14. |
| 5630. | +/- | 114. |
| 4.29 | +/- | 0. |
| 4.24 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.12 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
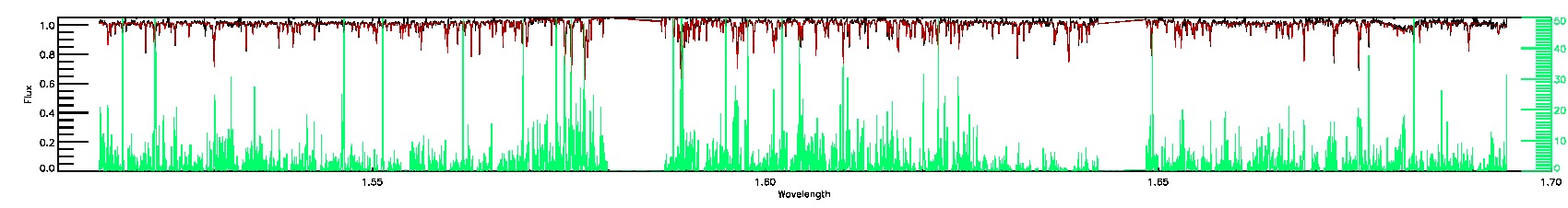
PERSIST_LOW
170-60
| 5857. | +/- | 12. |
| 5794. | +/- | 120. |
| 4.44 | +/- | 0. |
| 4.39 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.23 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.25 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
170-60
| 5338. | +/- | 21. |
| 5341. | +/- | 115. |
| 4.60 | +/- | 0. |
| 4.70 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 34.03 | +/- | 0. |
| 1.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.23 |
| -9999.00 |

PERSIST_LOW
170-60
| 4103. | +/- | 2. |
| 4202. | +/- | 72. |
| 4.43 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |

170-60
| 5588. | +/- | 12. |
| 5550. | +/- | 113. |
| 4.48 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.06 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.07 |
| -9999.00 |

170-60
| 4028. | +/- | 4. |
| 4126. | +/- | 80. |
| 4.46 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.11 | +/- | 0. |
| 7.77 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.02 |
| -9999.00 |

170-60
| 4783. | +/- | 4. |
| 4848. | +/- | 88. |
| 2.78 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.77 |
| -9999.00 |
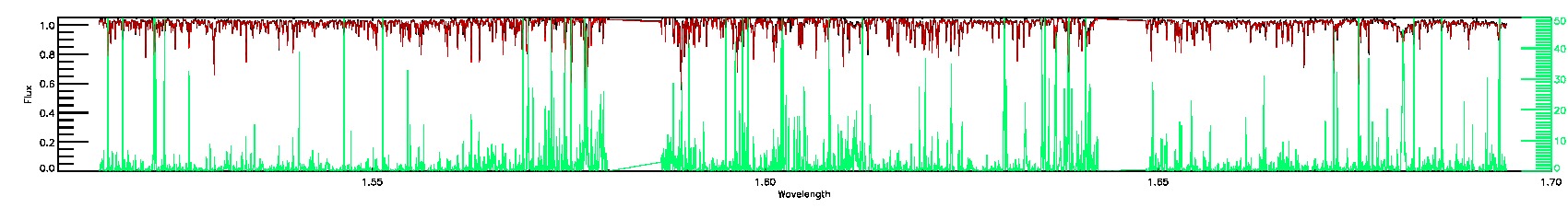
170-60
| 5109. | +/- | 6. |
| 5122. | +/- | 93. |
| 4.56 | +/- | 0. |
| 4.55 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.65 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
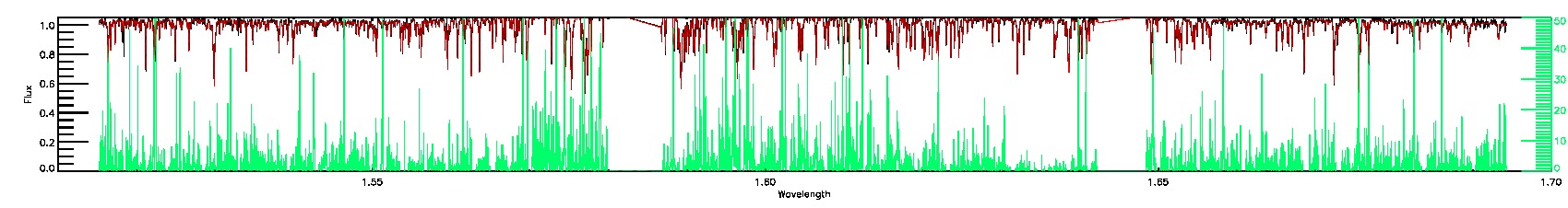
PERSIST_LOW
170-60
| 4956. | +/- | 9. |
| 4992. | +/- | 108. |
| 3.40 | +/- | 0. |
| 3.09 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.00 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.15 |
| -9999.00 |
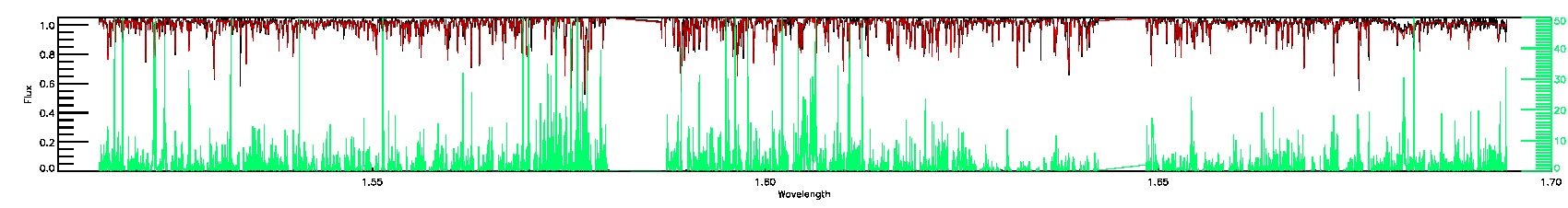
STAR_WARN,COLORTE_WARN
170-60
| 3458. | +/- | 3. |
| 3571. | +/- | 67. |
| 4.37 | +/- | 0. |
| 4.95 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -0.58 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 6.60 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.94 |
| -9999.00 |
170-60
| 5699. | +/- | 30. |
| 5652. | +/- | 146. |
| 4.47 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.77 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.12 |
| -9999.00 |
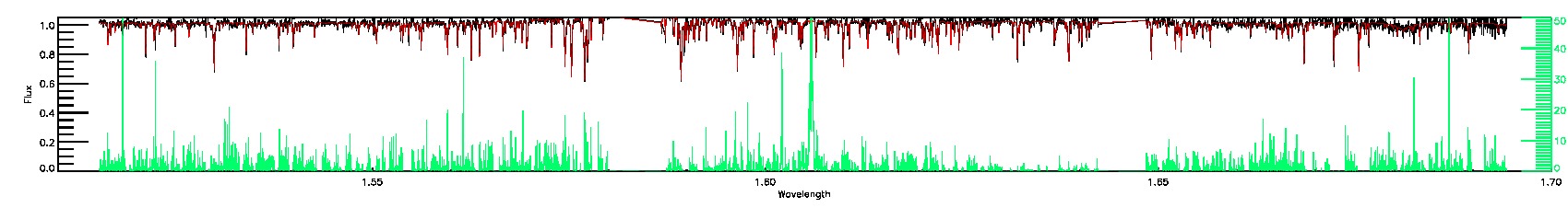
PERSIST_LOW
170-60
| 4662. | +/- | 4. |
| 4741. | +/- | 85. |
| 2.72 | +/- | 0. |
| 2.53 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.41 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
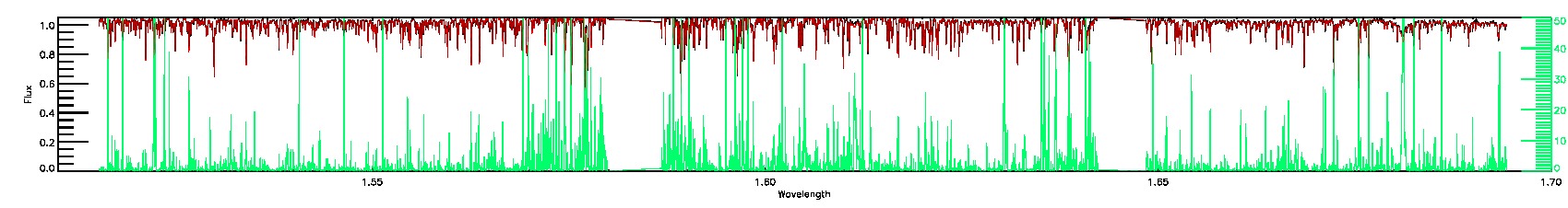
170-60
| 4016. | +/- | 2. |
| 4103. | +/- | 68. |
| 4.52 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.17 |
| -9999.00 |

170-60
| 6034. | +/- | 22. |
| 5948. | +/- | 141. |
| 4.38 | +/- | 0. |
| 4.27 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.00 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
BRIGHT_NEIGHBOR,PERSIST_LOW
170-60
| 5088. | +/- | 6. |
| 5112. | +/- | 94. |
| 2.91 | +/- | 0. |
| 2.54 | +/- | 0. |
| 1.79 | +/- | 0. |
| 1.79 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.09 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.09 |
| -9999.00 |

170-60
| 5232. | +/- | 10. |
| 5245. | +/- | 100. |
| 3.75 | +/- | 0. |
| 3.75 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.38 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 4418. | +/- | 4. |
| 4502. | +/- | 88. |
| 4.29 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.08 | +/- | 0. |
| 5.91 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.30 |
| -9999.00 |

170-60
| 5752. | +/- | 14. |
| 5691. | +/- | 120. |
| 4.51 | +/- | 0. |
| 4.38 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.53 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.19 |
| -9999.00 |

170-60
| 5395. | +/- | 10. |
| 5386. | +/- | 104. |
| 4.62 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.70 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
170-60
| 6397. | +/- | 17. |
| 6270. | +/- | 140. |
| 4.02 | +/- | 0. |
| 3.85 | +/- | 0. |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.11 | +/- | 0. |
| 53.22 | +/- | 0. |
| 1.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.19 |
| -9999.00 |
BRIGHT_NEIGHBOR
170-60
| 6132. | +/- | 23. |
| 6038. | +/- | 140. |
| 4.16 | +/- | 0. |
| 4.07 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 9.12 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.17 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
170-60
| 3480. | +/- | 4. |
| 3581. | +/- | 71. |
| 4.49 | +/- | 0. |
| 4.94 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.59 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.30 |
| -9999.00 |
PERSIST_LOW
170-60
| 5388. | +/- | 10. |
| 5374. | +/- | 120. |
| 4.61 | +/- | 0. |
| 4.60 | +/- | 0. |
| 1.60 | +/- | 0. |
| 1.60 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.81 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.08 |
| -9999.00 |

170-60
| 6386. | +/- | 18. |
| 6262. | +/- | 140. |
| 4.52 | +/- | 0. |
| 4.37 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 9.42 | +/- | 0. |
| 0.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.20 |
| -9999.00 |
170-60
| 5876. | +/- | 11. |
| 5796. | +/- | 135. |
| 4.19 | +/- | 0. |
| 3.99 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 3.59 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.54 |
| -9999.00 |

PERSIST_LOW,SUSPECT_BROAD_LINES
170-60
| 6801. | +/- | 17. |
| 6623. | +/- | 156. |
| 4.63 | +/- | 0. |
| 4.35 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 25.18 | +/- | 0. |
| 1.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.22 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
170-60
| 3467. | +/- | 2. |
| 3565. | +/- | 62. |
| 4.49 | +/- | 0. |
| 4.93 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 6.44 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.04 |
| -9999.00 |
170-60
| 6144. | +/- | 13. |
| 6035. | +/- | 126. |
| 4.20 | +/- | 0. |
| 3.97 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.62 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.47 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.32 |
| -9999.00 |
170-60
| 3776. | +/- | 2. |
| 3870. | +/- | 64. |
| 4.46 | +/- | 0. |
| 4.80 | +/- | 0. |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 4.15 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
170-60
| 6017. | +/- | 29. |
| 5952. | +/- | 143. |
| 4.40 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.86 | +/- | 0. |
| 0.86 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.17 | +/- | 0. |
| 5.55 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.10 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
170-60
| 3512. | +/- | 3. |
| 3605. | +/- | 69. |
| 4.41 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.57 | +/- | 0. |
| 0.57 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 5.03 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.77 |
| -9999.00 |
BRIGHT_NEIGHBOR
170-60
| 3811. | +/- | 6. |
| 3903. | +/- | 86. |
| 4.36 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 5.11 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
170-60
| 5115. | +/- | 9. |
| 5149. | +/- | 99. |
| 4.48 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.18 | +/- | 0. |
| 1.58 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
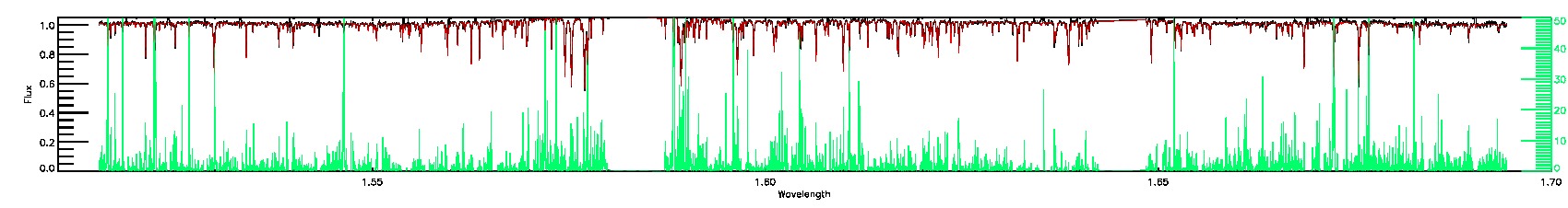
SUSPECT_BROAD_LINES
170-60
| 5990. | +/- | 14. |
| 5896. | +/- | 119. |
| 4.55 | +/- | 0. |
| 4.31 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 20.78 | +/- | 0. |
| 1.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.08 |
| -9999.00 |
BRIGHT_NEIGHBOR
170-60
| 5104. | +/- | 7. |
| 5117. | +/- | 104. |
| 4.65 | +/- | 0. |
| 4.63 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.85 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.28 |
| -9999.00 |

170-60
| 4974. | +/- | 18. |
| 5076. | +/- | 122. |
| 2.42 | +/- | 0. |
| 2.40 | +/- | 0. |
| 0.69 | +/- | 0. |
| 0.69 | +/- | -NaN |
| -1.60 | +/- | 0. |
| -1.60 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.54 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.17 | +/- | 0. |
| 7.52 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -2.16 |
| -9999.00 |
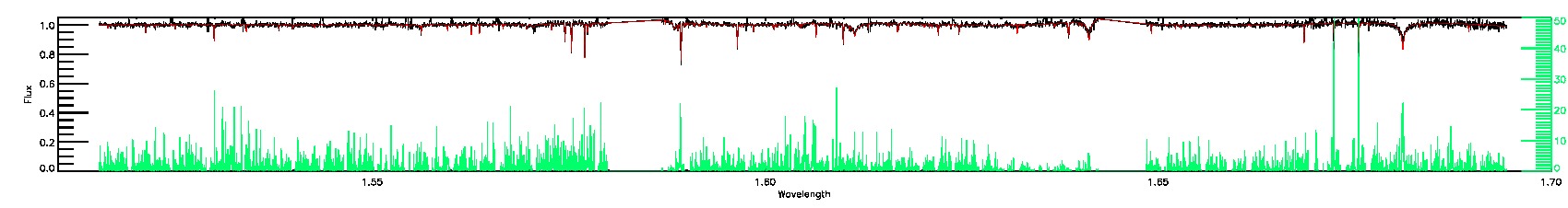
170-60
| 5768. | +/- | 11. |
| 5704. | +/- | 113. |
| 4.12 | +/- | 0. |
| 3.98 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 4.56 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
170-60
| 3424. | +/- | 3. |
| 3529. | +/- | 73. |
| 4.16 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 6.61 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
170-60
| 5816. | +/- | 16. |
| 5749. | +/- | 138. |
| 4.33 | +/- | 0. |
| 4.20 | +/- | 0. |
| 0.65 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.99 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.82 |
| -9999.00 |
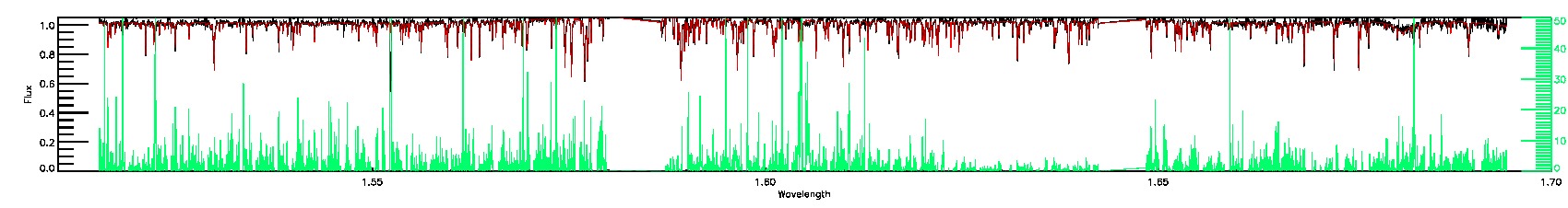
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,ROTATION_BAD
STAR_WARN,COLORTE_WARN,ROTATION_WARN
170-60
| 3600. | +/- | 6. |
| -10000. | +/- | -NaN |
| 3.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.73 | +/- | 0. |
| 1.73 | +/- | -NaN |
| -1.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.76 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.86 |
| -9999.00 |
170-60
| 5698. | +/- | 14. |
| 5650. | +/- | 116. |
| 4.40 | +/- | 0. |
| 4.34 | +/- | 0. |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.45 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.38 |
| -9999.00 |

170-60
| 5952. | +/- | 20. |
| 5879. | +/- | 138. |
| 4.28 | +/- | 0. |
| 4.22 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.32 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.34 |
| -9999.00 |
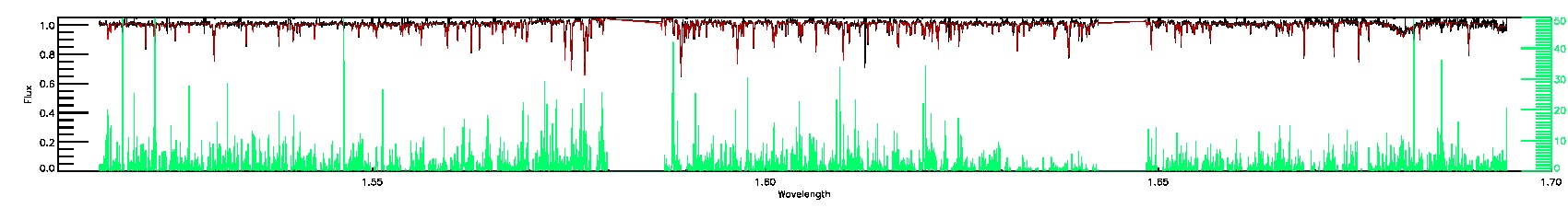
170-60
| 5552. | +/- | 14. |
| 5525. | +/- | 122. |
| 4.56 | +/- | 0. |
| 4.57 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.42 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |

170-60
| 5935. | +/- | 15. |
| 5856. | +/- | 138. |
| 4.37 | +/- | 0. |
| 4.24 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.42 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.15 |
| -9999.00 |

170-60
| 6076. | +/- | 17. |
| 5988. | +/- | 128. |
| 4.56 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.38 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.15 |
| -9999.00 |
PERSIST_LOW
170-60
| 5482. | +/- | 10. |
| 5461. | +/- | 107. |
| 4.47 | +/- | 0. |
| 4.48 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.64 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.10 |
| -9999.00 |

170-60
| 4881. | +/- | 6. |
| 4934. | +/- | 90. |
| 4.23 | +/- | 0. |
| 4.39 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.44 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
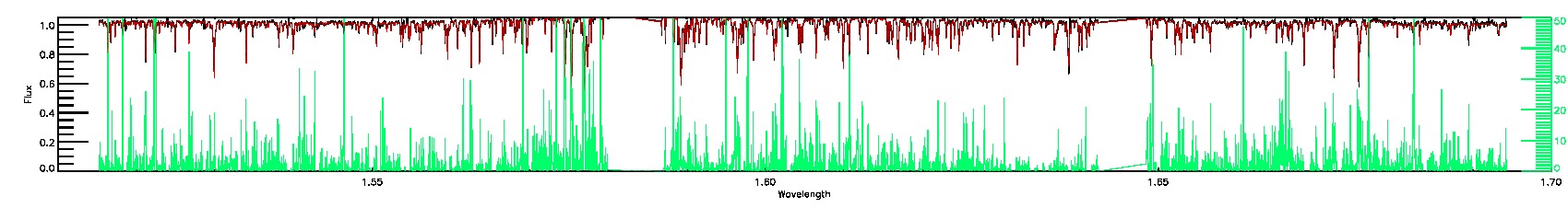
170-60
| 5114. | +/- | 14. |
| 5169. | +/- | 119. |
| 3.28 | +/- | 0. |
| 3.22 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.53 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -0.88 | +/- | 0. |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.17 | +/- | 0. |
| 4.95 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.70 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD
170-60
| 6540. | +/- | 20. |
| -10000. | +/- | -NaN |
| 3.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.98 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.43 |
| -9999.00 |
170-60
| 5273. | +/- | 9. |
| 5263. | +/- | 109. |
| 4.45 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.05 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_WARN,SN_WARN
170-60
| 5636. | +/- | 23. |
| 5593. | +/- | 147. |
| 4.56 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.06 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.16 |
| -9999.00 |

170-60
| 6270. | +/- | 17. |
| 6168. | +/- | 139. |
| 4.15 | +/- | 0. |
| 4.11 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.76 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 6.69 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
170-60
| 5149. | +/- | 8. |
| 5168. | +/- | 97. |
| 4.63 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.13 | +/- | 0. |
| 8.00 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |

PERSIST_LOW
170-60
| 4346. | +/- | 2. |
| 4440. | +/- | 81. |
| 4.40 | +/- | 0. |
| 4.64 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.34 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.13 |
| -9999.00 |

170-60
| 3819. | +/- | 3. |
| 3914. | +/- | 77. |
| 4.46 | +/- | 0. |
| 4.80 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.98 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
PERSIST_LOW
170-60
| 4717. | +/- | 8. |
| 4793. | +/- | 105. |
| 3.97 | +/- | 0. |
| 4.19 | +/- | 0. |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 12.74 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.09 |
| -9999.00 |
170-60
| 4409. | +/- | 2. |
| 4503. | +/- | 78. |
| 4.43 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.60 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |

170-60
| 5801. | +/- | 22. |
| 5746. | +/- | 155. |
| 4.55 | +/- | 0. |
| 4.52 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.19 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 1.94 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.43 |
| -9999.00 |
170-60
| 5663. | +/- | 12. |
| 5620. | +/- | 112. |
| 4.46 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.53 | +/- | 0. |
| 0.53 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.64 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
PERSIST_LOW
170-60
| 5445. | +/- | 13. |
| 5415. | +/- | 128. |
| 4.56 | +/- | 0. |
| 4.44 | +/- | 0. |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.21 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 1.62 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.49 |
| -9999.00 |
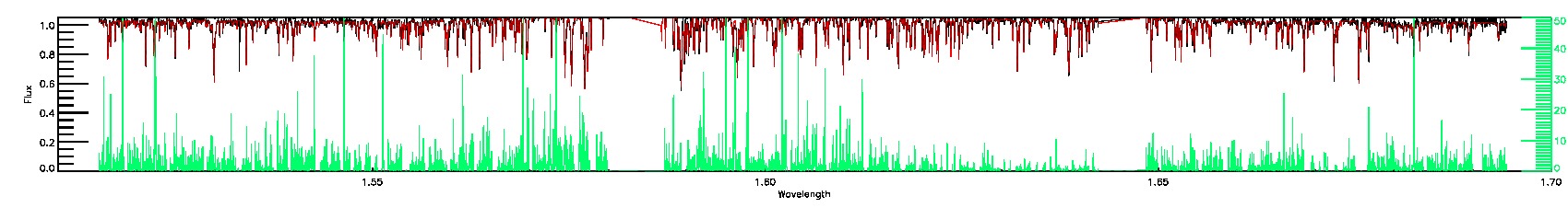
PERSIST_LOW
170-60
| 5405. | +/- | 8. |
| 5384. | +/- | 113. |
| 4.50 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.59 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.01 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.01 |
| -9999.00 |

170-60
| 5125. | +/- | 17. |
| 5163. | +/- | 131. |
| 4.46 | +/- | 0. |
| 4.70 | +/- | 0. |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -0.53 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 2.07 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.60 |
| -9999.00 |

170-60
| 4892. | +/- | 9. |
| 4933. | +/- | 105. |
| 4.60 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.60 | +/- | 0. |
| 1.60 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.50 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.35 |
| -9999.00 |

170-60
| 6246. | +/- | 16. |
| 6132. | +/- | 132. |
| 4.34 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 17.36 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
170-60
| 5747. | +/- | 12. |
| 5684. | +/- | 114. |
| 4.52 | +/- | 0. |
| 4.37 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.85 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.23 |
| -9999.00 |

PERSIST_LOW
170-60
| 5619. | +/- | 12. |
| 5581. | +/- | 122. |
| 4.20 | +/- | 0. |
| 4.17 | +/- | 0. |
| 0.47 | +/- | 0. |
| 0.47 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.11 | +/- | 0. |
| 1.60 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.07 |
| -9999.00 |

170-60
| 5148. | +/- | 12. |
| 5158. | +/- | 113. |
| 3.86 | +/- | 0. |
| 3.81 | +/- | 0. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.82 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.99 |
| -9999.00 |

170-60
| 5386. | +/- | 11. |
| 5381. | +/- | 105. |
| 3.70 | +/- | 0. |
| 3.66 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 10.61 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
170-60
| 3224. | +/- | 2. |
| 3320. | +/- | 55. |
| 4.67 | +/- | 0. |
| 5.14 | +/- | 0. |
| 2.72 | +/- | 0. |
| 2.72 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 14.02 | +/- | 0. |
| 1.15 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
170-60
| 4712. | +/- | 7. |
| 4767. | +/- | 97. |
| 4.27 | +/- | 0. |
| 4.29 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.22 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.08 | +/- | 0. |
| 4.37 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
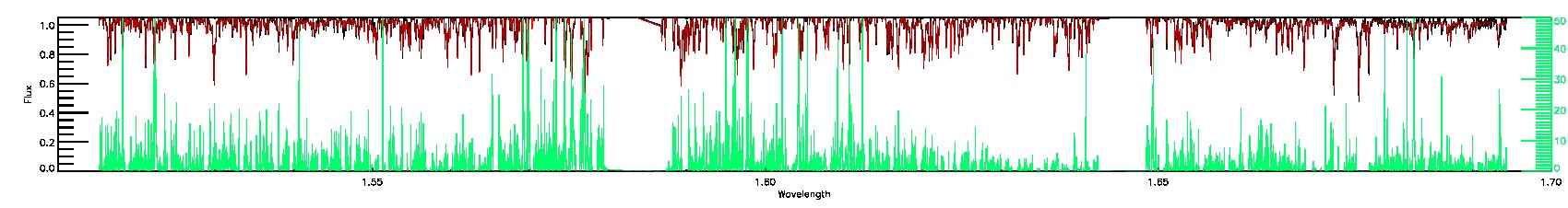
170-60
| 3886. | +/- | 1. |
| 3996. | +/- | 70. |
| 1.14 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.77 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.09 | +/- | 0. |
| 3.97 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.62 |
| -9999.00 |
170-60
| 5396. | +/- | 17. |
| 5396. | +/- | 118. |
| 4.48 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.87 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.44 |
| -9999.00 |

170-60
| 5448. | +/- | 17. |
| 5430. | +/- | 128. |
| 4.35 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.57 | +/- | 0. |
| 0.57 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.96 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
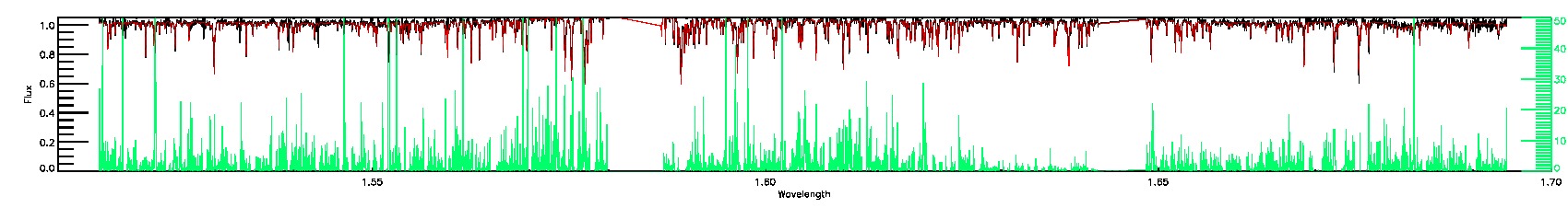
170-60
| 4601. | +/- | 4. |
| 4697. | +/- | 86. |
| 4.39 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.66 | +/- | 0. |
| 0.66 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.46 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.29 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.30 |
| -9999.00 |

170-60
| 5958. | +/- | 29. |
| 5888. | +/- | 152. |
| 4.31 | +/- | 0. |
| 4.29 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.61 |
| -9999.00 |
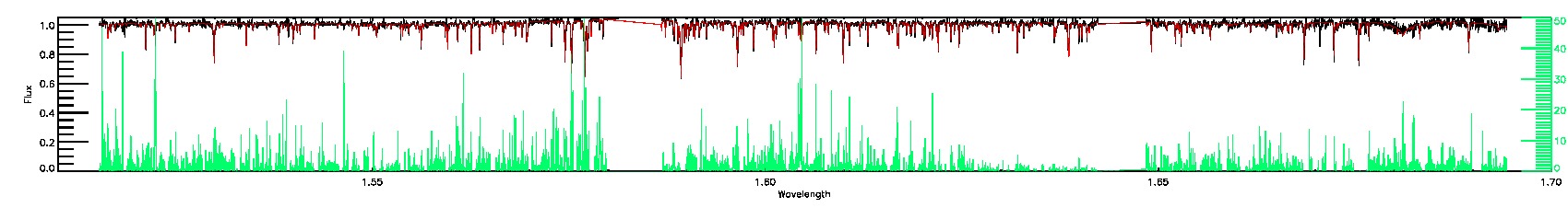
BRIGHT_NEIGHBOR
170-60
| 6001. | +/- | 25. |
| 5927. | +/- | 145. |
| 4.25 | +/- | 0. |
| 4.23 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.54 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.19 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
170-60
| 8438. | +/- | 43. |
| 8293. | +/- | 328. |
| 4.35 | +/- | 0. |
| 4.47 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 76.37 | +/- | 0. |
| 1.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.20 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -1.31 |
| -9999.00 |
170-60
| 5137. | +/- | 19. |
| 5245. | +/- | 127. |
| 2.92 | +/- | 0. |
| 3.01 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -2.19 | +/- | 0. |
| -2.19 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.03 | +/- | 1. |
| -0.03 | +/- | -NaN |
| 0.41 | +/- | 0. |
| 0.38 | +/- | 0. |
| 10.66 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.35 |
| -9999.00 |
170-60
| 5954. | +/- | 18. |
| 5883. | +/- | 136. |
| 4.21 | +/- | 0. |
| 4.17 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.02 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
170-60
| 5783. | +/- | 15. |
| 5729. | +/- | 135. |
| 4.47 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.54 | +/- | 0. |
| 0.54 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.72 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.91 |
| -9999.00 |

170-60
| 5661. | +/- | 26. |
| 5616. | +/- | 140. |
| 4.47 | +/- | 0. |
| 4.41 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.84 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
170-60
| 5581. | +/- | 10. |
| 5537. | +/- | 110. |
| 4.37 | +/- | 0. |
| 4.25 | +/- | 0. |
| 0.48 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.08 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
170-60
| 3592. | +/- | 5. |
| 3681. | +/- | 75. |
| 4.66 | +/- | 0. |
| 4.98 | +/- | 0. |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 1.57 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
BRIGHT_NEIGHBOR
170-60
| 5885. | +/- | 16. |
| 5820. | +/- | 134. |
| 4.17 | +/- | 0. |
| 4.13 | +/- | 0. |
| 0.86 | +/- | 0. |
| 0.86 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.74 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
170-60
| 5073. | +/- | 21. |
| 5132. | +/- | 132. |
| 3.15 | +/- | 0. |
| 2.78 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -0.88 | +/- | 0. |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.23 | +/- | 0. |
| 4.94 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.40 |
| -9999.00 |

170-60
| 4185. | +/- | 1. |
| 4284. | +/- | 74. |
| 1.98 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.41 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.44 |
| -9999.00 |

BRIGHT_NEIGHBOR
170-60
| 5058. | +/- | 8. |
| 5081. | +/- | 106. |
| 4.53 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 1.62 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |