STAR_WARN,COLORTE_WARN
002-03-O
| 8787. | +/- | 43. |
| 8688. | +/- | 371. |
| 4.07 | +/- | 0. |
| 4.62 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 26.36 | +/- | 0. |
| 1.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.33 |
| -9999.00 |
002-03-O
| 8304. | +/- | 44. |
| 8123. | +/- | 298. |
| 4.23 | +/- | 0. |
| 4.01 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 79.45 | +/- | 0. |
| 1.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.44 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.90 |
| -9999.00 |
002-03-O
| 8067. | +/- | 25. |
| 7922. | +/- | 265. |
| 4.02 | +/- | 0. |
| 4.15 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 87.20 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.17 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.51 |
| -9999.00 |
STAR_BAD
STAR_WARN,SN_WARN
002-03-O
| 5906. | +/- | 50. |
| -10000. | +/- | -NaN |
| 2.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.46 | +/- | 1. |
| 4.46 | +/- | -NaN |
| -2.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.72 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.03 |
| -9999.00 |

STAR_WARN,SN_WARN
002-03-O
| 4480. | +/- | 17. |
| 4565. | +/- | 112. |
| 2.44 | +/- | 0. |
| 2.26 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.83 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.98 |
| -9999.00 |

002-03-O
| 8284. | +/- | 30. |
| 8175. | +/- | 311. |
| 4.50 | +/- | 0. |
| 4.97 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 59.50 | +/- | 0. |
| 1.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -2.16 |
| -9999.00 |
002-03-O
| 8587. | +/- | 40. |
| 8488. | +/- | 350. |
| 3.96 | +/- | 0. |
| 4.52 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 43.51 | +/- | 0. |
| 1.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.38 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 4350. | +/- | 11. |
| -10000. | +/- | -NaN |
| 2.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.36 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.58 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
002-03-O
| 8244. | +/- | 22. |
| 8144. | +/- | 315. |
| 4.17 | +/- | 0. |
| 4.73 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.35 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.38 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.29 |
| -9999.00 |
STAR_WARN,SN_WARN
002-03-O
| 7446. | +/- | 41. |
| 7218. | +/- | 245. |
| 3.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.21 | +/- | 0. |
| 4.21 | +/- | -NaN |
| 0.60 | +/- | 0. |
| 0.60 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 8.39 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.98 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD
002-03-O
| 8500. | +/- | 29. |
| -10000. | +/- | -NaN |
| 4.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.64 | +/- | 0. |
| 1.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.36 |
| -9999.00 |
002-03-O
| 8509. | +/- | 28. |
| 8411. | +/- | 343. |
| 4.26 | +/- | 0. |
| 4.84 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.40 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.42 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
002-03-O
| 4608. | +/- | 30. |
| 4712. | +/- | 126. |
| 2.21 | +/- | 0. |
| 2.06 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -0.66 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.77 |
| -9999.00 |
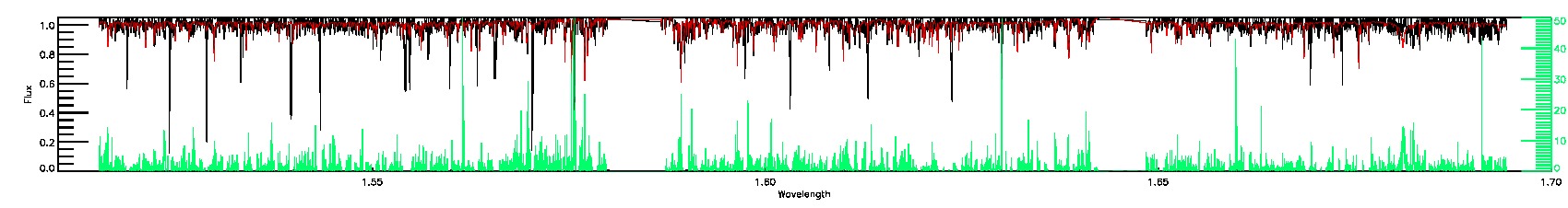
SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
002-03-O
| 8532. | +/- | 44. |
| 8412. | +/- | 349. |
| 4.38 | +/- | 0. |
| 4.74 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 41.39 | +/- | 0. |
| 1.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.13 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.01 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
002-03-O
| 8277. | +/- | 23. |
| 8179. | +/- | 320. |
| 4.35 | +/- | 0. |
| 4.93 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.62 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.36 |
| -9999.00 |
002-03-O
| 8509. | +/- | 31. |
| 8405. | +/- | 337. |
| 3.94 | +/- | 0. |
| 4.46 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 31.87 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.42 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.09 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 4439. | +/- | 24. |
| -10000. | +/- | -NaN |
| 2.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.71 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.46 |
| -9999.00 |
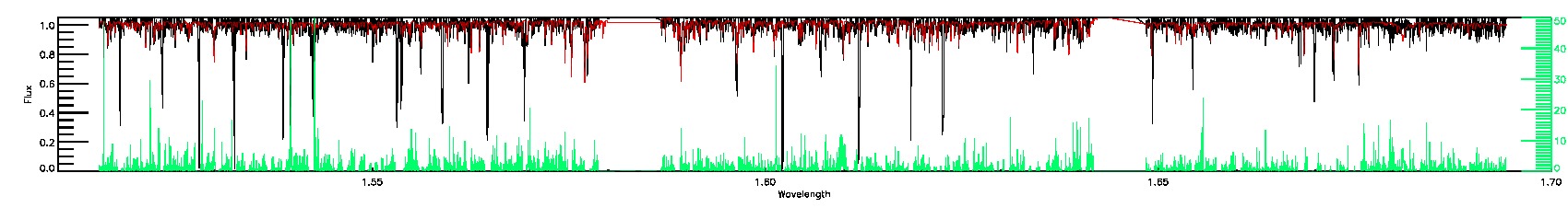
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_WARN,SN_WARN
002-03-O
| 6151. | +/- | 88. |
| 6049. | +/- | 185. |
| 3.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.40 | +/- | 0. |
| 2.40 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.04 | +/- | 1. |
| -0.04 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.15 | +/- | 0. |
| 35.58 | +/- | 0. |
| 1.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
002-03-O
| 6608. | +/- | 42. |
| -10000. | +/- | -NaN |
| 5.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.32 | +/- | 1. |
| 2.32 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.63 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.43 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
002-03-O
| 4429. | +/- | 11. |
| 4510. | +/- | 107. |
| 2.54 | +/- | 0. |
| 2.29 | +/- | 0. |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.05 | +/- | 0. |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.88 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
002-03-O
| 7074. | +/- | 69. |
| -10000. | +/- | -NaN |
| 3.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -1.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.55 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.49 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
002-03-O
| 11997. | +/- | 63. |
| -10000. | +/- | -NaN |
| 4.36 | +/- | 0. |
| 4.21 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.27 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.62 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.75 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
002-03-O
| 8884. | +/- | 54. |
| 8735. | +/- | 334. |
| 3.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 80.69 | +/- | 0. |
| 1.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -1.09 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5305. | +/- | 131. |
| -10000. | +/- | -NaN |
| 2.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.42 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.25 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.89 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5617. | +/- | 114. |
| -10000. | +/- | -NaN |
| 2.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.67 | +/- | 0. |
| 4.67 | +/- | -NaN |
| -1.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.47 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.15 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.20 |
| -9999.00 |
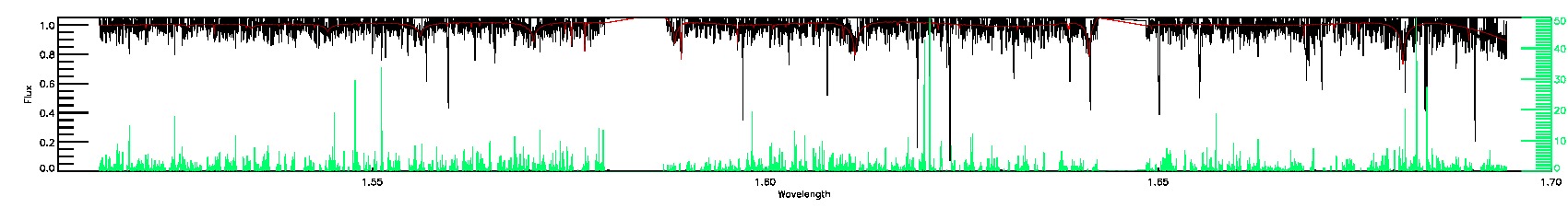
STAR_WARN,COLORTE_WARN
002-03-O
| 13185. | +/- | 96. |
| -10000. | +/- | -NaN |
| 4.48 | +/- | 0. |
| 4.18 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.72 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.51 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
002-03-O
| 4283. | +/- | 21. |
| -10000. | +/- | -NaN |
| 2.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.95 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.53 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4892. | +/- | 141. |
| -10000. | +/- | -NaN |
| 2.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.37 | +/- | 1. |
| 0.37 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.62 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.66 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.98 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 5191. | +/- | 81. |
| -10000. | +/- | -NaN |
| 3.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.69 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.52 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7796. | +/- | 173. |
| -10000. | +/- | -NaN |
| 3.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.67 | +/- | 0. |
| 4.67 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.13 | +/- | 1. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.20 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5811. | +/- | 100. |
| -10000. | +/- | -NaN |
| 2.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.59 | +/- | 0. |
| 4.59 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.29 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 1.45 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
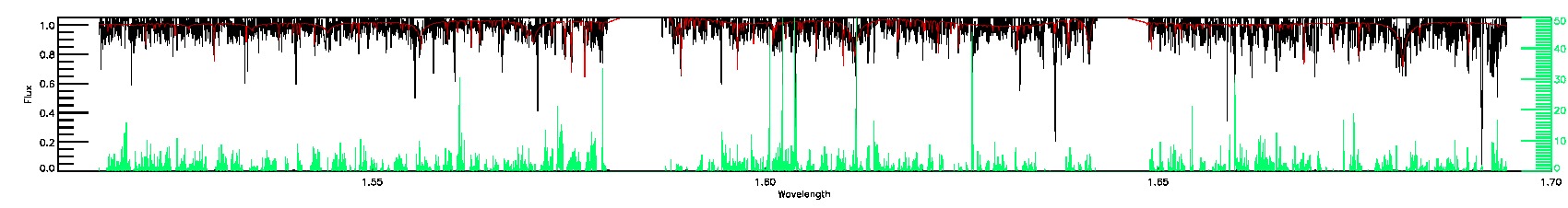
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 6852. | +/- | 69. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.62 | +/- | 1. |
| 1.62 | +/- | -NaN |
| -1.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.78 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 83.20 | +/- | 0. |
| 1.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.40 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5756. | +/- | 119. |
| -10000. | +/- | -NaN |
| 2.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.51 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.91 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.48 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 5312. | +/- | 119. |
| -10000. | +/- | -NaN |
| 2.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -0.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.29 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.07 |
| -9999.00 |
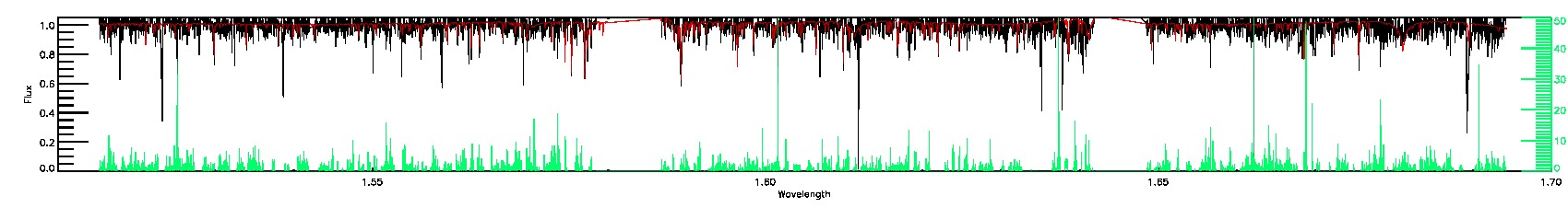
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7942. | +/- | 152. |
| -10000. | +/- | -NaN |
| 2.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.23 | +/- | 0. |
| 1.23 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 25.69 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5588. | +/- | 140. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.53 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.57 |
| -9999.00 |
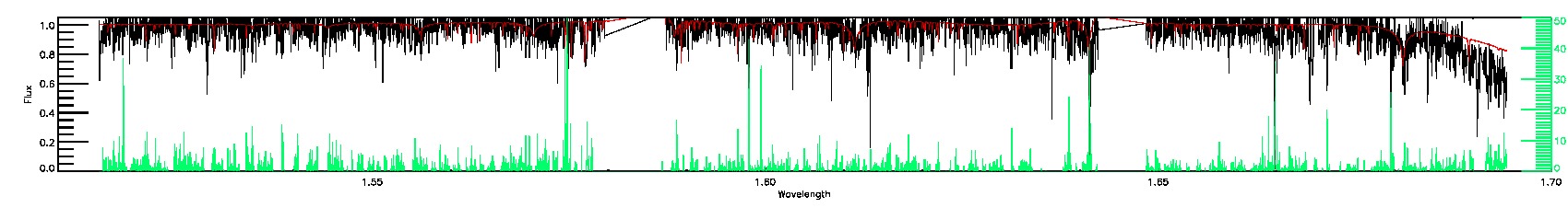
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7653. | +/- | 104. |
| -10000. | +/- | -NaN |
| 4.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.93 | +/- | 1. |
| 1.93 | +/- | -NaN |
| -1.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.04 | +/- | 1. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.30 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5670. | +/- | 147. |
| -10000. | +/- | -NaN |
| 2.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.55 | +/- | 0. |
| 4.55 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.01 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.42 |
| -9999.00 |

STAR_WARN,SN_WARN
002-03-O
| 4488. | +/- | 14. |
| 4567. | +/- | 109. |
| 2.53 | +/- | 0. |
| 2.29 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.61 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7383. | +/- | 139. |
| -10000. | +/- | -NaN |
| 3.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.06 | +/- | 0. |
| 4.06 | +/- | -NaN |
| -0.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.02 | +/- | 1. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.92 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5685. | +/- | 123. |
| -10000. | +/- | -NaN |
| 2.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 2. |
| 0.97 | +/- | -NaN |
| -2.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 181. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 89. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.88 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.90 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.87 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6585. | +/- | 125. |
| -10000. | +/- | -NaN |
| 2.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.47 | +/- | 0. |
| 2.47 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 59.29 | +/- | 0. |
| 1.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5879. | +/- | 106. |
| -10000. | +/- | -NaN |
| 2.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.05 | +/- | 1. |
| 1.05 | +/- | -NaN |
| -1.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.59 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 11. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.39 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.92 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.98 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.47 |
| -9999.00 |
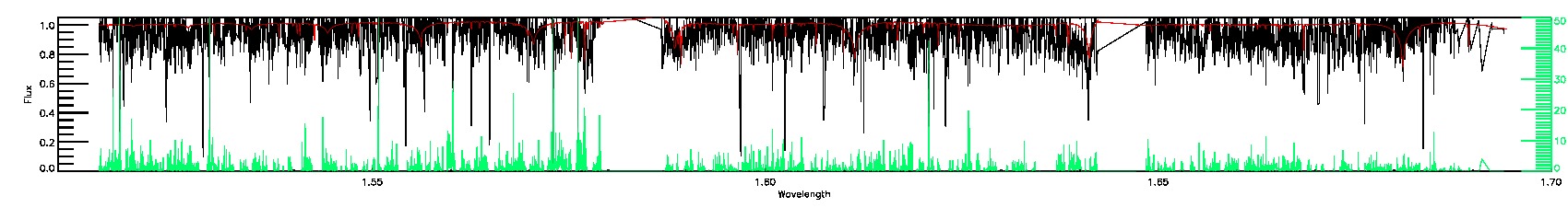
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6630. | +/- | 103. |
| -10000. | +/- | -NaN |
| 2.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.77 | +/- | 0. |
| 4.77 | +/- | -NaN |
| -1.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.01 | +/- | 1. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.31 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7745. | +/- | 225. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.46 | +/- | 1. |
| 1.46 | +/- | -NaN |
| -0.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.92 | +/- | 1. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.51 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3821. | +/- | 38. |
| -10000. | +/- | -NaN |
| 0.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.48 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.95 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4371. | +/- | 107. |
| -10000. | +/- | -NaN |
| 1.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| -1.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.92 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.79 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.56 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 1.13 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4510. | +/- | 51. |
| -10000. | +/- | -NaN |
| 2.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.61 | +/- | 0. |
| 0.61 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.31 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.09 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7957. | +/- | 203. |
| -10000. | +/- | -NaN |
| 2.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.38 | +/- | 0. |
| 0.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5535. | +/- | 54. |
| -10000. | +/- | -NaN |
| 3.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.90 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 8307. | +/- | 485. |
| -10000. | +/- | -NaN |
| 3.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.53 | +/- | 3. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.96 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.30 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5915. | +/- | 105. |
| -10000. | +/- | -NaN |
| 2.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.65 | +/- | 2. |
| 0.65 | +/- | -NaN |
| -2.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.53 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.10 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7908. | +/- | 176. |
| -10000. | +/- | -NaN |
| 3.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.12 | +/- | 1. |
| 1.12 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 20.18 | +/- | 0. |
| 1.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.15 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5220. | +/- | 121. |
| -10000. | +/- | -NaN |
| 1.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.60 | +/- | 0. |
| 4.60 | +/- | -NaN |
| -1.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.74 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 1.37 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -0.93 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 6403. | +/- | 38. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.43 | +/- | 0. |
| 4.43 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 18.95 | +/- | 0. |
| 1.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.79 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7604. | +/- | 136. |
| -10000. | +/- | -NaN |
| 3.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.61 | +/- | 1. |
| 4.61 | +/- | -NaN |
| -1.75 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.92 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 27.49 | +/- | 0. |
| 1.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5337. | +/- | 142. |
| -10000. | +/- | -NaN |
| 2.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| -0.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.11 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5288. | +/- | 108. |
| -10000. | +/- | -NaN |
| 1.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.73 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.99 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6293. | +/- | 131. |
| -10000. | +/- | -NaN |
| 2.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.14 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.35 |
| -9999.00 |
STAR_WARN,SN_WARN
002-03-O
| 3388. | +/- | 6. |
| 3480. | +/- | 82. |
| 0.56 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.09 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.37 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5330. | +/- | 181. |
| -10000. | +/- | -NaN |
| 3.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.74 | +/- | 2. |
| 0.74 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.00 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.48 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7139. | +/- | 77. |
| -10000. | +/- | -NaN |
| 3.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.78 | +/- | 0. |
| 4.78 | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 56.70 | +/- | 0. |
| 1.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.03 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5044. | +/- | 114. |
| -10000. | +/- | -NaN |
| 1.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.57 | +/- | 0. |
| 0.57 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.46 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -2.10 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 5500. | +/- | 0. |
| -10000. | +/- | -NaN |
| 3.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 24.87 | +/- | 0. |
| 1.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.39 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5306. | +/- | 80. |
| -10000. | +/- | -NaN |
| 2.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.60 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.94 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.81 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4935. | +/- | 110. |
| -10000. | +/- | -NaN |
| 1.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| -1.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.37 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.61 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6441. | +/- | 40. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.26 | +/- | 1. |
| 1.26 | +/- | -NaN |
| -0.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 39.67 | +/- | 0. |
| 1.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5364. | +/- | 79. |
| -10000. | +/- | -NaN |
| 1.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| -1.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.58 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.54 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5141. | +/- | 91. |
| -10000. | +/- | -NaN |
| 1.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.41 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.59 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.18 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7391. | +/- | 157. |
| -10000. | +/- | -NaN |
| 3.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.91 | +/- | 5. |
| 0.91 | +/- | -NaN |
| -2.12 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 13.15 | +/- | 1. |
| 1.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -1.95 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 11671. | +/- | 398. |
| -10000. | +/- | -NaN |
| 3.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.02 | +/- | 2. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.83 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6072. | +/- | 147. |
| -10000. | +/- | -NaN |
| 3.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 21.84 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.37 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6536. | +/- | 118. |
| -10000. | +/- | -NaN |
| 2.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.41 | +/- | 0. |
| 4.41 | +/- | -NaN |
| -1.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 22.45 | +/- | 0. |
| 1.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.53 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 6202. | +/- | 108. |
| -10000. | +/- | -NaN |
| 2.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.79 | +/- | 0. |
| 4.79 | +/- | -NaN |
| -1.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.26 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -2.48 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5903. | +/- | 114. |
| -10000. | +/- | -NaN |
| 2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.02 | +/- | 0. |
| 2.02 | +/- | -NaN |
| -0.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.23 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.29 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 6734. | +/- | 90. |
| -10000. | +/- | -NaN |
| 2.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.04 | +/- | 0. |
| 1.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.95 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6074. | +/- | 70. |
| -10000. | +/- | -NaN |
| 2.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.74 | +/- | 0. |
| 1.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.21 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5089. | +/- | 97. |
| -10000. | +/- | -NaN |
| 1.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| -0.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.05 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.50 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7485. | +/- | 151. |
| -10000. | +/- | -NaN |
| 3.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.05 | +/- | 1. |
| 2.05 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.71 | +/- | 0. |
| 1.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.43 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 4153. | +/- | 25. |
| -10000. | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.30 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.86 |
| -9999.00 |
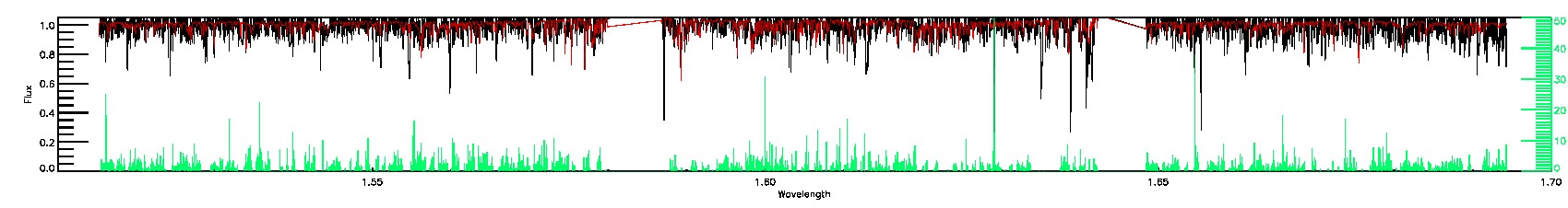
002-03-O
| 3243. | +/- | 2. |
| 3325. | +/- | 67. |
| 0.51 | +/- | 0. |
| 0.33 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 2.70 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.32 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7870. | +/- | 162. |
| -10000. | +/- | -NaN |
| 3.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.40 | +/- | 0. |
| 4.40 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 35.05 | +/- | 0. |
| 1.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.85 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7488. | +/- | 155. |
| -10000. | +/- | -NaN |
| 3.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 14.89 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.46 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7380. | +/- | 203. |
| -10000. | +/- | -NaN |
| 3.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.98 | +/- | 5. |
| 1.98 | +/- | -NaN |
| -2.36 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 3.16 | +/- | 6. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.35 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7943. | +/- | 150. |
| -10000. | +/- | -NaN |
| 4.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.80 | +/- | 1. |
| 0.80 | +/- | -NaN |
| 0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.96 | +/- | 0. |
| 1.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.98 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5899. | +/- | 113. |
| -10000. | +/- | -NaN |
| 2.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.19 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.39 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3684. | +/- | 26. |
| -10000. | +/- | -NaN |
| 1.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.91 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -2.35 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5746. | +/- | 63. |
| -10000. | +/- | -NaN |
| 2.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.12 | +/- | 0. |
| 1.12 | +/- | -NaN |
| -0.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.04 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6889. | +/- | 189. |
| -10000. | +/- | -NaN |
| 2.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.79 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7910. | +/- | 149. |
| -10000. | +/- | -NaN |
| 3.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.93 | +/- | 0. |
| 1.93 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 9. |
| -9999.99 | +/- | -NaN |
| 0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.40 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.50 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6545. | +/- | 86. |
| -10000. | +/- | -NaN |
| 2.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 91.83 | +/- | 0. |
| 1.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.19 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4410. | +/- | 110. |
| -10000. | +/- | -NaN |
| 3.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.80 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -2.32 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 6190. | +/- | 76. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.92 | +/- | 1. |
| 0.92 | +/- | -NaN |
| -1.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 93.63 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5348. | +/- | 101. |
| -10000. | +/- | -NaN |
| 1.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.12 | +/- | 1. |
| 1.12 | +/- | -NaN |
| -1.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.83 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |

STAR_BAD
STAR_WARN,SN_WARN
002-03-O
| 5820. | +/- | 54. |
| -10000. | +/- | -NaN |
| 2.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.68 | +/- | 0. |
| 4.68 | +/- | -NaN |
| -1.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.60 | +/- | 0. |
| 0.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.45 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.12 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4912. | +/- | 64. |
| -10000. | +/- | -NaN |
| 1.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.15 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5832. | +/- | 115. |
| -10000. | +/- | -NaN |
| 2.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.96 | +/- | 0. |
| 2.96 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.51 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7145. | +/- | 162. |
| -10000. | +/- | -NaN |
| 3.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.64 | +/- | 0. |
| 2.64 | +/- | -NaN |
| -0.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.65 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 18.06 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 1.48 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.16 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5552. | +/- | 93. |
| -10000. | +/- | -NaN |
| 1.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.48 | +/- | 1. |
| 1.48 | +/- | -NaN |
| -1.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.22 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.06 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5888. | +/- | 111. |
| -10000. | +/- | -NaN |
| 2.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 0. |
| 0.53 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.19 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.80 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5673. | +/- | 55. |
| -10000. | +/- | -NaN |
| 2.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 24.55 | +/- | 0. |
| 1.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.06 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5946. | +/- | 116. |
| -10000. | +/- | -NaN |
| 2.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.09 | +/- | 1. |
| 1.09 | +/- | -NaN |
| -0.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 20.65 | +/- | 0. |
| 1.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.44 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4491. | +/- | 120. |
| -10000. | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.29 | +/- | 0. |
| 2.29 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.93 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.81 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 12.72 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.84 |
| -9999.00 |
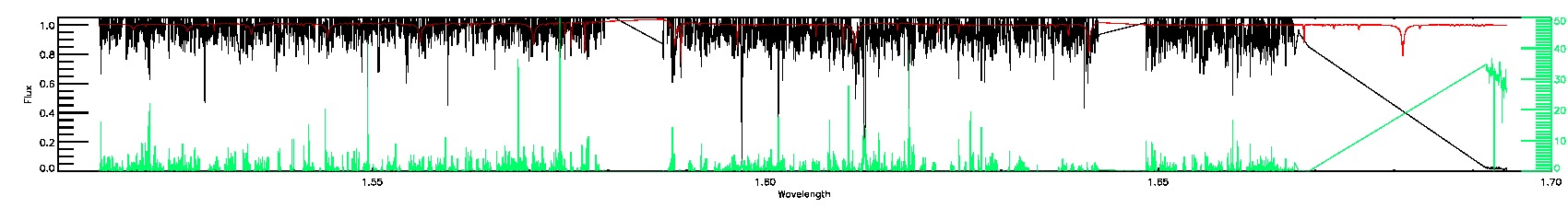
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5182. | +/- | 131. |
| -10000. | +/- | -NaN |
| 1.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -1.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.62 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -2.49 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 7385. | +/- | 89. |
| -10000. | +/- | -NaN |
| 3.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.62 | +/- | 0. |
| 4.62 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.02 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.68 |
| -9999.00 |
STAR_WARN,SN_WARN
002-03-O
| 3935. | +/- | 8. |
| 4036. | +/- | 99. |
| 1.54 | +/- | 0. |
| 1.38 | +/- | 0. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.53 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.26 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4267. | +/- | 38. |
| -10000. | +/- | -NaN |
| 1.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.35 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.63 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5749. | +/- | 0. |
| -10000. | +/- | -NaN |
| 2.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.20 | +/- | 0. |
| 1.20 | +/- | -NaN |
| -1.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.60 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.86 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 4815. | +/- | 54. |
| -10000. | +/- | -NaN |
| 1.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -1.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.60 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 1.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 3117. | +/- | 37. |
| -10000. | +/- | -NaN |
| 2.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.87 | +/- | 0. |
| 0.87 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.78 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 2.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.34 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6416. | +/- | 80. |
| -10000. | +/- | -NaN |
| 2.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.08 | +/- | 0. |
| 2.08 | +/- | -NaN |
| -1.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.63 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6429. | +/- | 111. |
| -10000. | +/- | -NaN |
| 2.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 40.16 | +/- | 0. |
| 1.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -2.45 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6128. | +/- | 0. |
| -10000. | +/- | -NaN |
| 2.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| 1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 14.00 | +/- | 0. |
| 1.15 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.52 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 6682. | +/- | 92. |
| -10000. | +/- | -NaN |
| 2.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.60 | +/- | 0. |
| 3.60 | +/- | -NaN |
| -1.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 12. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.25 | +/- | 1. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.67 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7359. | +/- | 127. |
| -10000. | +/- | -NaN |
| 3.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 0. |
| 0.98 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.40 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5648. | +/- | 96. |
| -10000. | +/- | -NaN |
| 2.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.82 | +/- | 1. |
| 0.82 | +/- | -NaN |
| -1.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.68 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.23 |
| -9999.00 |

STAR_WARN,SN_WARN
002-03-O
| 3374. | +/- | 2. |
| 3459. | +/- | 74. |
| 0.56 | +/- | 0. |
| 0.40 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.82 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.14 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6145. | +/- | 136. |
| -10000. | +/- | -NaN |
| 2.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.53 | +/- | 0. |
| 4.53 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.63 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.41 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7869. | +/- | 160. |
| -10000. | +/- | -NaN |
| 3.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 44.90 | +/- | 0. |
| 1.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.19 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5702. | +/- | 129. |
| -10000. | +/- | -NaN |
| 2.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.42 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
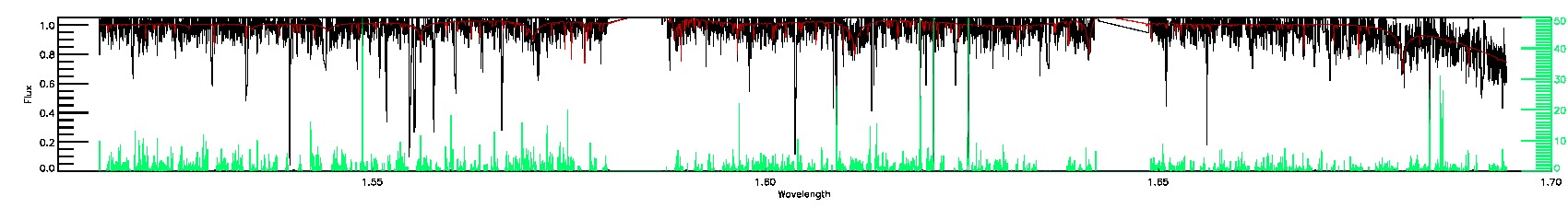
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6220. | +/- | 147. |
| -10000. | +/- | -NaN |
| 2.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.91 | +/- | 0. |
| 1.14 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.58 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4353. | +/- | 1. |
| -10000. | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.74 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.90 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 1.95 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.01 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7105. | +/- | 147. |
| -10000. | +/- | -NaN |
| 2.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.38 | +/- | 1. |
| 1.38 | +/- | -NaN |
| -1.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 85.55 | +/- | 0. |
| 1.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 0.22 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7250. | +/- | 128. |
| -10000. | +/- | -NaN |
| 3.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.14 | +/- | 0. |
| 3.14 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.16 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.03 |
| -9999.00 |
STAR_WARN,SN_WARN
002-03-O
| 3693. | +/- | 5. |
| 3776. | +/- | 87. |
| 1.38 | +/- | 0. |
| 1.19 | +/- | 0. |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.75 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.65 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6557. | +/- | 207. |
| -10000. | +/- | -NaN |
| 2.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.91 | +/- | 2. |
| 0.91 | +/- | -NaN |
| -1.26 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 82.62 | +/- | 0. |
| 1.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.42 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5662. | +/- | 112. |
| -10000. | +/- | -NaN |
| 1.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.89 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.91 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4970. | +/- | 100. |
| -10000. | +/- | -NaN |
| 1.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.60 | +/- | 0. |
| 1.60 | +/- | -NaN |
| -2.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.24 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.37 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.45 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6898. | +/- | 172. |
| -10000. | +/- | -NaN |
| 2.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| 0.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 89.58 | +/- | 0. |
| 1.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 1.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.36 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3079. | +/- | 41. |
| -10000. | +/- | -NaN |
| 4.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.84 | +/- | 0. |
| 2.84 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.79 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.22 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.83 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6205. | +/- | 94. |
| -10000. | +/- | -NaN |
| 2.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.79 | +/- | 0. |
| 2.79 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 35.88 | +/- | 0. |
| 1.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.04 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5336. | +/- | 112. |
| -10000. | +/- | -NaN |
| 1.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.61 | +/- | 1. |
| 4.61 | +/- | -NaN |
| -2.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.70 | +/- | 13. |
| -9999.99 | +/- | -NaN |
| 0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.81 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.36 |
| -9999.00 |
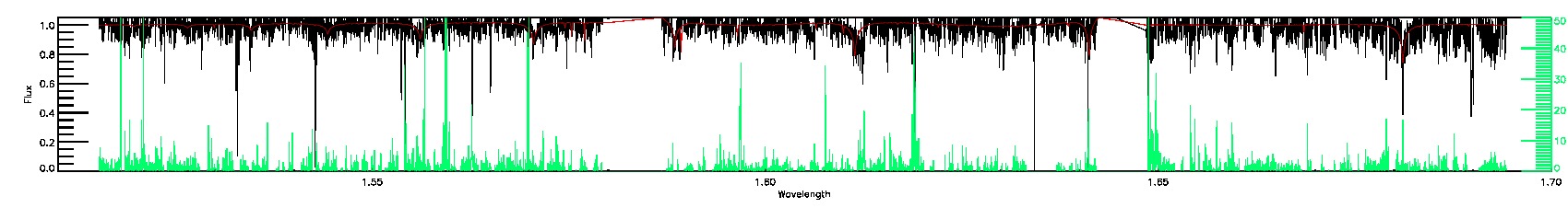
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7751. | +/- | 155. |
| -10000. | +/- | -NaN |
| 3.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 18.07 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.45 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6802. | +/- | 148. |
| -10000. | +/- | -NaN |
| 2.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.61 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 66.02 | +/- | 0. |
| 1.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5712. | +/- | 104. |
| -10000. | +/- | -NaN |
| 2.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| -1.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.42 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.41 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 3650. | +/- | 6. |
| -10000. | +/- | -NaN |
| 0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| -0.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.21 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.73 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6256. | +/- | 94. |
| -10000. | +/- | -NaN |
| 2.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.76 | +/- | 0. |
| 1.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 1.49 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.81 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 4634. | +/- | 56. |
| -10000. | +/- | -NaN |
| 1.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.15 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.10 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7780. | +/- | 174. |
| -10000. | +/- | -NaN |
| 4.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.73 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.14 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6076. | +/- | 57. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.95 | +/- | 1. |
| 0.95 | +/- | -NaN |
| -0.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.38 | +/- | 0. |
| 0.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -1.25 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7962. | +/- | 162. |
| -10000. | +/- | -NaN |
| 3.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 26.04 | +/- | 0. |
| 1.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.87 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6185. | +/- | 63. |
| -10000. | +/- | -NaN |
| 2.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.75 | +/- | 0. |
| 1.75 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.36 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.38 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5925. | +/- | 130. |
| -10000. | +/- | -NaN |
| 2.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.06 | +/- | 2. |
| 1.06 | +/- | -NaN |
| -1.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.74 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.59 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 1.03 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |

002-03-O
| 3018. | +/- | 1. |
| 3130. | +/- | 58. |
| 0.77 | +/- | 0. |
| 0.69 | +/- | 0. |
| 3.21 | +/- | 0. |
| 3.21 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.08 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.47 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.55 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7072. | +/- | 124. |
| -10000. | +/- | -NaN |
| 3.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 42.35 | +/- | 0. |
| 1.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.14 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6479. | +/- | 97. |
| -10000. | +/- | -NaN |
| 2.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.58 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.86 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6334. | +/- | 16. |
| -10000. | +/- | -NaN |
| 3.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.73 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.38 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5596. | +/- | 81. |
| -10000. | +/- | -NaN |
| 2.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.41 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.74 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5388. | +/- | 114. |
| -10000. | +/- | -NaN |
| 1.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 1. |
| 0.55 | +/- | -NaN |
| -1.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.46 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7920. | +/- | 167. |
| -10000. | +/- | -NaN |
| 3.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.87 | +/- | 1. |
| 0.87 | +/- | -NaN |
| 0.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 28.81 | +/- | 0. |
| 1.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| 0.88 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7292. | +/- | 201. |
| -10000. | +/- | -NaN |
| 3.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 88.04 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.11 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4123. | +/- | 50. |
| -10000. | +/- | -NaN |
| 0.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -1.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.34 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.80 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3277. | +/- | 10. |
| -10000. | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.81 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.48 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 3418. | +/- | 8. |
| -10000. | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.05 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.53 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6197. | +/- | 105. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.15 | +/- | 0. |
| 1.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5639. | +/- | 93. |
| -10000. | +/- | -NaN |
| 2.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.07 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 1.34 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6183. | +/- | 67. |
| -10000. | +/- | -NaN |
| 2.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.70 | +/- | 43. |
| 0.70 | +/- | -NaN |
| -2.40 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 68. |
| -9999.99 | +/- | -NaN |
| -0.65 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 94.67 | +/- | 1. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6959. | +/- | 103. |
| -10000. | +/- | -NaN |
| 3.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.92 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7599. | +/- | 222. |
| -10000. | +/- | -NaN |
| 4.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.40 | +/- | 0. |
| 2.40 | +/- | -NaN |
| 0.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 93.00 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.83 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5808. | +/- | 92. |
| -10000. | +/- | -NaN |
| 2.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.62 | +/- | 1. |
| 0.62 | +/- | -NaN |
| -1.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 14.92 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.19 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7422. | +/- | 210. |
| -10000. | +/- | -NaN |
| 3.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 1. |
| 0.48 | +/- | -NaN |
| 0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 94.45 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.10 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5806. | +/- | 117. |
| -10000. | +/- | -NaN |
| 3.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 27.54 | +/- | 0. |
| 1.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 1.47 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5785. | +/- | 110. |
| -10000. | +/- | -NaN |
| 2.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.86 | +/- | 1. |
| 0.86 | +/- | -NaN |
| -1.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.37 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6433. | +/- | 83. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -1.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.40 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.27 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
002-03-O
| 3448. | +/- | 10. |
| -10000. | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -1.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.07 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.24 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4276. | +/- | 54. |
| -10000. | +/- | -NaN |
| 0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.87 | +/- | 0. |
| 1.87 | +/- | -NaN |
| -2.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.63 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.25 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.79 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5696. | +/- | 134. |
| -10000. | +/- | -NaN |
| 2.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.95 | +/- | 0. |
| 0.95 | +/- | -NaN |
| -1.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.36 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.27 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5910. | +/- | 28. |
| -10000. | +/- | -NaN |
| 1.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -1.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.93 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| 0.99 |
| -9999.00 |
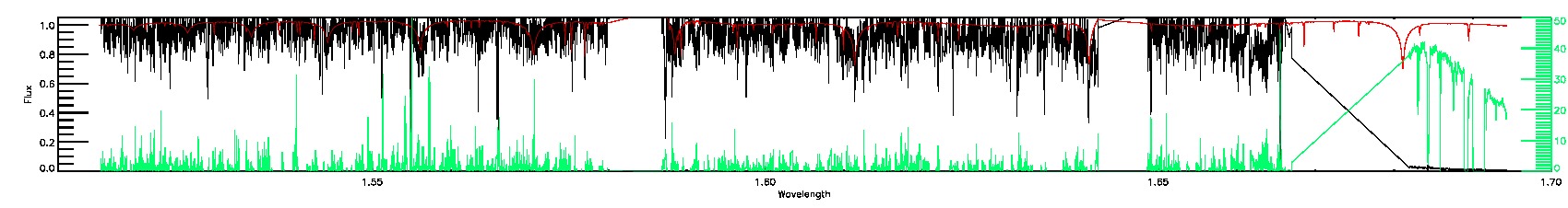
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6790. | +/- | 89. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.51 | +/- | 0. |
| 1.51 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 88.72 | +/- | 0. |
| 1.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.17 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4625. | +/- | 71. |
| -10000. | +/- | -NaN |
| 0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -1.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.59 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3887. | +/- | 24. |
| -10000. | +/- | -NaN |
| 1.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.33 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.67 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6222. | +/- | 132. |
| -10000. | +/- | -NaN |
| 2.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.07 | +/- | 1. |
| 1.07 | +/- | -NaN |
| -0.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.86 | +/- | 0. |
| 1.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.80 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4591. | +/- | 75. |
| -10000. | +/- | -NaN |
| 0.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.19 | +/- | 1. |
| 1.19 | +/- | -NaN |
| -2.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.93 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.95 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.93 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.31 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7605. | +/- | 162. |
| -10000. | +/- | -NaN |
| 3.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.84 | +/- | 2. |
| 1.84 | +/- | -NaN |
| -1.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.22 | +/- | 1. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.87 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5758. | +/- | 106. |
| -10000. | +/- | -NaN |
| 1.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.60 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 1.67 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.48 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6528. | +/- | 176. |
| -10000. | +/- | -NaN |
| 3.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.42 | +/- | 0. |
| 1.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
STAR_WARN,SN_WARN
002-03-O
| 3711. | +/- | 15. |
| 3879. | +/- | 110. |
| 0.68 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -1.85 | +/- | 0. |
| -1.85 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.25 | +/- | 0. |
| 8.74 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.47 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7892. | +/- | 169. |
| -10000. | +/- | -NaN |
| 3.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.92 | +/- | 1. |
| 0.92 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.28 | +/- | 0. |
| 0.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_BAD
STAR_WARN,SN_WARN
002-03-O
| 3034. | +/- | 2. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.45 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.99 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4878. | +/- | 0. |
| -10000. | +/- | -NaN |
| 5.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 25.90 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -2.18 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5852. | +/- | 95. |
| -10000. | +/- | -NaN |
| 2.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.36 | +/- | 1. |
| 1.36 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.83 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -0.31 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7344. | +/- | 191. |
| -10000. | +/- | -NaN |
| 2.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 81.88 | +/- | 0. |
| 1.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3900. | +/- | 3. |
| -10000. | +/- | -NaN |
| 0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -2.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.59 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.34 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5828. | +/- | 105. |
| -10000. | +/- | -NaN |
| 2.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| -0.80 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.72 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.45 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7943. | +/- | 147. |
| -10000. | +/- | -NaN |
| 3.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.06 | +/- | 1. |
| 1.06 | +/- | -NaN |
| -0.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.46 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.84 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4123. | +/- | 6. |
| -10000. | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.75 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.28 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 1.57 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
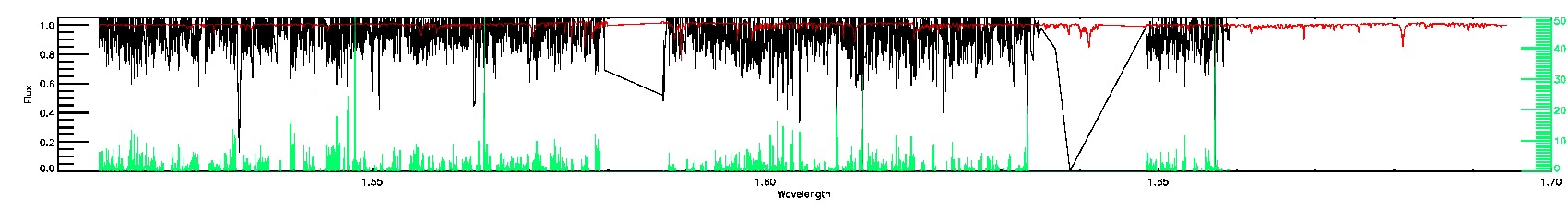
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7935. | +/- | 230. |
| -10000. | +/- | -NaN |
| 3.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.05 | +/- | 0. |
| 3.05 | +/- | -NaN |
| 0.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 69.81 | +/- | 0. |
| 1.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5579. | +/- | 107. |
| -10000. | +/- | -NaN |
| 1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.45 | +/- | 1. |
| 2.45 | +/- | -NaN |
| -1.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.87 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.55 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -1.52 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7842. | +/- | 154. |
| -10000. | +/- | -NaN |
| 3.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.85 | +/- | 0. |
| 1.85 | +/- | -NaN |
| 0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.19 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.42 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.98 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7257. | +/- | 159. |
| -10000. | +/- | -NaN |
| 3.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.81 | +/- | 3. |
| 0.81 | +/- | -NaN |
| -2.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 23.42 | +/- | 1. |
| 1.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.39 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 3933. | +/- | 4. |
| -10000. | +/- | -NaN |
| 1.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -1.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.39 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.73 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 1.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.57 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 4962. | +/- | 156. |
| -10000. | +/- | -NaN |
| 2.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.14 | +/- | 1. |
| 1.14 | +/- | -NaN |
| -1.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.41 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.99 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7912. | +/- | 183. |
| -10000. | +/- | -NaN |
| 3.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.12 | +/- | 1. |
| 3.12 | +/- | -NaN |
| -1.16 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 42.69 | +/- | 0. |
| 1.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.91 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 4189. | +/- | 41. |
| -10000. | +/- | -NaN |
| 4.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.87 | +/- | 0. |
| 0.87 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 32.89 | +/- | 0. |
| 1.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.47 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 1.50 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.35 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5247. | +/- | 136. |
| -10000. | +/- | -NaN |
| 1.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.54 | +/- | 1. |
| 0.54 | +/- | -NaN |
| -1.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.26 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.96 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6239. | +/- | 128. |
| -10000. | +/- | -NaN |
| 2.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 2. |
| 0.98 | +/- | -NaN |
| -1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 25.17 | +/- | 0. |
| 1.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 7074. | +/- | 208. |
| -10000. | +/- | -NaN |
| 3.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.77 | +/- | 1. |
| 0.77 | +/- | -NaN |
| 0.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 44.26 | +/- | 0. |
| 1.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.95 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3309. | +/- | 9. |
| -10000. | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.04 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 3208. | +/- | 23. |
| -10000. | +/- | -NaN |
| 2.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.63 | +/- | 0. |
| 0.63 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.83 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 2.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.29 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 5308. | +/- | 130. |
| -10000. | +/- | -NaN |
| 1.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.02 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 2.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.93 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
002-03-O
| 3025. | +/- | 4. |
| -10000. | +/- | -NaN |
| 1.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| 1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.73 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6155. | +/- | 0. |
| -10000. | +/- | -NaN |
| 2.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| 0.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.41 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.45 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 1.50 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 5806. | +/- | 151. |
| -10000. | +/- | -NaN |
| 2.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.89 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.84 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
002-03-O
| 6235. | +/- | 137. |
| -10000. | +/- | -NaN |
| 2.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.72 | +/- | 0. |
| 1.72 | +/- | -NaN |
| -0.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.10 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.71 |
| -9999.00 |
