005-03-O
| 8364. | +/- | 28. |
| 8235. | +/- | 301. |
| 4.17 | +/- | 0. |
| 4.44 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 47.38 | +/- | 0. |
| 1.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.01 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.68 |
| -9999.00 |
STAR_WARN,SN_WARN
005-03-O
| 3861. | +/- | 5. |
| 3946. | +/- | 93. |
| 1.64 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 2.83 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5172. | +/- | 79. |
| -10000. | +/- | -NaN |
| 1.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.80 | +/- | 0. |
| 4.80 | +/- | -NaN |
| -1.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.75 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.39 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
005-03-O
| 8585. | +/- | 54. |
| 8483. | +/- | 362. |
| 3.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 49.87 | +/- | 0. |
| 1.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.38 |
| -9999.00 |
005-03-O
| 3297. | +/- | 1. |
| 3370. | +/- | 53. |
| 0.70 | +/- | 0. |
| 0.50 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| 0.37 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.41 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 6639. | +/- | 119. |
| -10000. | +/- | -NaN |
| 3.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.79 | +/- | 0. |
| 1.79 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 38.21 | +/- | 0. |
| 1.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.76 |
| -9999.00 |
005-03-O
| 3646. | +/- | 1. |
| 3744. | +/- | 63. |
| 0.79 | +/- | 0. |
| 0.67 | +/- | 0. |
| 1.90 | +/- | 0. |
| 1.90 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.11 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.28 |
| -9999.00 |
005-03-O
| 4564. | +/- | 4. |
| 4633. | +/- | 89. |
| 2.78 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.53 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.19 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD
STAR_WARN,COLORTE_WARN
005-03-O
| 7996. | +/- | 11. |
| -10000. | +/- | -NaN |
| 4.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.81 | +/- | 0. |
| 3.81 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 44.19 | +/- | 0. |
| 1.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5993. | +/- | 102. |
| -10000. | +/- | -NaN |
| 2.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 1. |
| 0.96 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.42 | +/- | 0. |
| 1.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -1.92 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 6537. | +/- | 60. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.19 | +/- | 1. |
| 1.19 | +/- | -NaN |
| -1.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 6. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 80.89 | +/- | 0. |
| 1.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.42 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
005-03-O
| 8934. | +/- | 58. |
| -10000. | +/- | -NaN |
| 3.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.08 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.59 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.53 |
| -9999.00 |
SUSPECT_RV_COMBINATION
005-03-O
| 8647. | +/- | 38. |
| 8548. | +/- | 356. |
| 3.88 | +/- | 0. |
| 4.44 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 21.36 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.08 |
| -9999.00 |
SUSPECT_RV_COMBINATION
005-03-O
| 8450. | +/- | 27. |
| 8351. | +/- | 336. |
| 3.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.20 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.10 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.23 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 6523. | +/- | 61. |
| -10000. | +/- | -NaN |
| 2.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.84 | +/- | 0. |
| 2.84 | +/- | -NaN |
| -1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.38 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.49 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.73 |
| -9999.00 |
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
005-03-O
| 6180. | +/- | 26. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.56 | +/- | 0. |
| 4.56 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.57 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |
005-03-O
| 4027. | +/- | 2. |
| 4120. | +/- | 85. |
| 1.80 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.13 | +/- | 0. |
| 3.13 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.42 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 4546. | +/- | 31. |
| -10000. | +/- | -NaN |
| 2.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.02 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.46 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 7070. | +/- | 87. |
| -10000. | +/- | -NaN |
| 2.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.50 | +/- | 1. |
| 1.50 | +/- | -NaN |
| -2.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 11. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 23.86 | +/- | 0. |
| 1.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.14 |
| -9999.00 |
005-03-O
| 4420. | +/- | 4. |
| 4492. | +/- | 94. |
| 2.87 | +/- | 0. |
| 2.61 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.40 | +/- | 0. |
| 0.40 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.34 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.88 |
| -9999.00 |
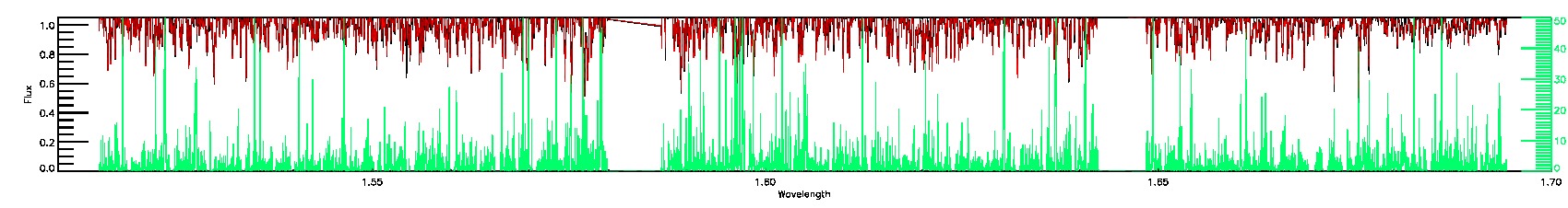
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD
STAR_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5372. | +/- | 47. |
| -10000. | +/- | -NaN |
| 1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.08 | +/- | 0. |
| 3.08 | +/- | -NaN |
| -2.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.93 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 22. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.42 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.82 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.30 |
| -9999.00 |

STAR_WARN,SN_WARN
005-03-O
| 3998. | +/- | 3. |
| 4089. | +/- | 91. |
| 1.69 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.07 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.27 |
| -9999.00 |

005-03-O
| 3871. | +/- | 2. |
| 3951. | +/- | 80. |
| 1.49 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.63 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.57 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
005-03-O
| 6041. | +/- | 31. |
| -10000. | +/- | -NaN |
| 4.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.77 | +/- | 0. |
| 4.77 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.83 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.76 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
005-03-O
| 8678. | +/- | 41. |
| 8549. | +/- | 332. |
| 3.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 74.46 | +/- | 0. |
| 1.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.38 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -1.72 |
| -9999.00 |
STAR_WARN,SN_WARN
005-03-O
| 4600. | +/- | 11. |
| 4667. | +/- | 108. |
| 2.83 | +/- | 0. |
| 2.48 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.58 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.46 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 4671. | +/- | 56. |
| -10000. | +/- | -NaN |
| 1.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
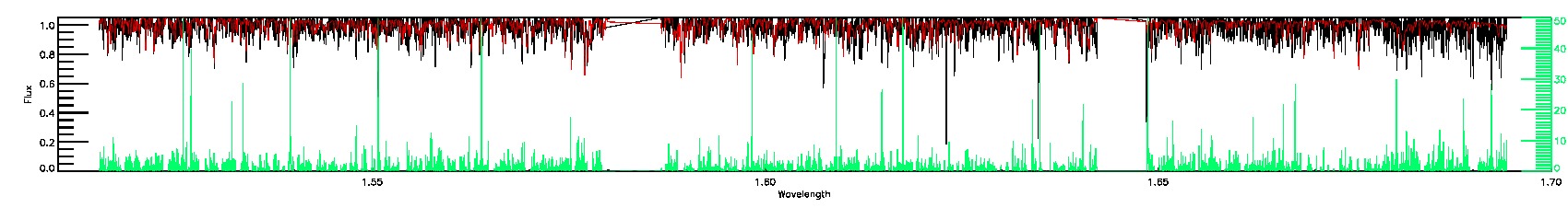
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 6218. | +/- | 98. |
| -10000. | +/- | -NaN |
| 3.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 32.24 | +/- | 0. |
| 1.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
005-03-O
| 7863. | +/- | 13. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| -1.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.85 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.31 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
005-03-O
| 8569. | +/- | 31. |
| 8452. | +/- | 332. |
| 4.24 | +/- | 0. |
| 4.64 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.47 | +/- | 0. |
| 0.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -1.73 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN
005-03-O
| 8460. | +/- | 27. |
| 8359. | +/- | 335. |
| 3.94 | +/- | 0. |
| 4.48 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.95 | +/- | 0. |
| 1.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.37 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.31 |
| -9999.00 |
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
005-03-O
| 4591. | +/- | 37. |
| -10000. | +/- | -NaN |
| 1.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.06 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.92 |
| -9999.00 |

005-03-O
| 3994. | +/- | 2. |
| 4129. | +/- | 88. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | 0. |
| 2.23 | +/- | 0. |
| 2.23 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -1.08 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.26 | +/- | 0. |
| 5.57 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.78 |
| -9999.00 |

STAR_WARN,SN_WARN
005-03-O
| 4004. | +/- | 3. |
| 4071. | +/- | 86. |
| 2.02 | +/- | 0. |
| 1.72 | +/- | 0. |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| 0.49 | +/- | 0. |
| 0.49 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.22 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.84 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5900. | +/- | 56. |
| -10000. | +/- | -NaN |
| 2.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.82 | +/- | 1. |
| 0.82 | +/- | -NaN |
| -1.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 24.20 | +/- | 0. |
| 1.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
STAR_BAD
STAR_WARN,SN_WARN
005-03-O
| 7466. | +/- | 57. |
| -10000. | +/- | -NaN |
| 3.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.53 | +/- | 0. |
| 4.53 | +/- | -NaN |
| -0.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.24 | +/- | 0. |
| 1.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.99 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6294. | +/- | 60. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.73 | +/- | 0. |
| 1.73 | +/- | -NaN |
| -1.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 44.15 | +/- | 0. |
| 1.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.77 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6777. | +/- | 130. |
| -10000. | +/- | -NaN |
| 3.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.12 | +/- | 0. |
| 4.12 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.98 | +/- | 0. |
| 1.30 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -2.02 |
| -9999.00 |
BRIGHT_NEIGHBOR,LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6991. | +/- | 105. |
| -10000. | +/- | -NaN |
| 4.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.42 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_WARN,SN_WARN
005-03-O
| 3908. | +/- | 4. |
| 3988. | +/- | 89. |
| 1.55 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.63 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
005-03-O
| 6268. | +/- | 24. |
| -10000. | +/- | -NaN |
| 3.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.89 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.37 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5381. | +/- | 119. |
| -10000. | +/- | -NaN |
| 1.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.28 | +/- | 1. |
| 1.28 | +/- | -NaN |
| -2.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 6. |
| -9999.99 | +/- | -NaN |
| 0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.63 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -2.48 |
| -9999.00 |

BRIGHT_NEIGHBOR,LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7845. | +/- | 164. |
| -10000. | +/- | -NaN |
| 3.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.64 | +/- | 1. |
| 1.64 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.59 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.71 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
005-03-O
| 4083. | +/- | 2. |
| 4201. | +/- | 94. |
| 1.65 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -0.69 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.23 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.96 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
005-03-O
| 4278. | +/- | 4. |
| 4411. | +/- | 104. |
| 1.69 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.76 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.28 | +/- | 0. |
| 5.48 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.67 |
| -9999.00 |
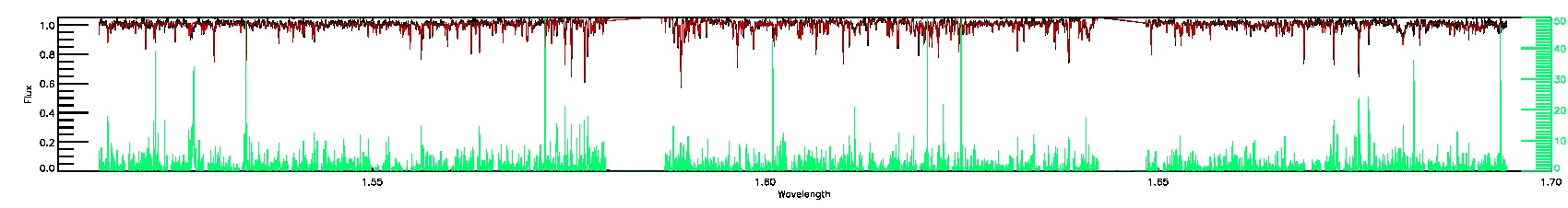
SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
005-03-O
| 8858. | +/- | 43. |
| 8760. | +/- | 380. |
| 3.97 | +/- | 0. |
| 4.55 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.65 | +/- | 1. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.38 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.27 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6354. | +/- | 125. |
| -10000. | +/- | -NaN |
| 2.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.69 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 31.54 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.54 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
005-03-O
| 8575. | +/- | 48. |
| 8473. | +/- | 382. |
| 4.02 | +/- | 0. |
| 4.55 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -2.33 | +/- | 0. |
| -2.33 | +/- | 1. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 2. |
| 18.23 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -2.23 |
| -9999.00 |
005-03-O
| 3691. | +/- | 1. |
| 3766. | +/- | 71. |
| 1.35 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.45 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
STAR_WARN,SN_WARN
005-03-O
| 3985. | +/- | 3. |
| 4054. | +/- | 86. |
| 1.92 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.46 | +/- | 0. |
| 0.46 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.26 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.73 |
| -9999.00 |
STAR_WARN,SN_WARN
005-03-O
| 5690. | +/- | 33. |
| 5715. | +/- | 186. |
| 2.31 | +/- | 0. |
| 2.31 | +/- | 0. |
| 3.37 | +/- | 0. |
| 3.37 | +/- | -NaN |
| -1.75 | +/- | 0. |
| -1.75 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.09 | +/- | 3. |
| 0.09 | +/- | -NaN |
| 0.42 | +/- | 0. |
| 0.39 | +/- | 0. |
| 8.23 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.36 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5571. | +/- | 76. |
| -10000. | +/- | -NaN |
| 2.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.85 | +/- | 0. |
| 1.85 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 88.78 | +/- | 0. |
| 1.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6816. | +/- | 111. |
| -10000. | +/- | -NaN |
| 3.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.55 | +/- | 1. |
| 1.55 | +/- | -NaN |
| -1.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 6. |
| -9999.99 | +/- | -NaN |
| 0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.64 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.46 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_WARN,SN_WARN
005-03-O
| 4544. | +/- | 17. |
| 4633. | +/- | 115. |
| 2.42 | +/- | 0. |
| 2.25 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.17 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.13 |
| -9999.00 |

LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5674. | +/- | 57. |
| -10000. | +/- | -NaN |
| 2.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.24 | +/- | 0. |
| 2.24 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 25.94 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.94 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5449. | +/- | 125. |
| -10000. | +/- | -NaN |
| 1.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.57 | +/- | 2. |
| 0.57 | +/- | -NaN |
| -2.43 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 15. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 12.27 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.46 |
| -9999.00 |
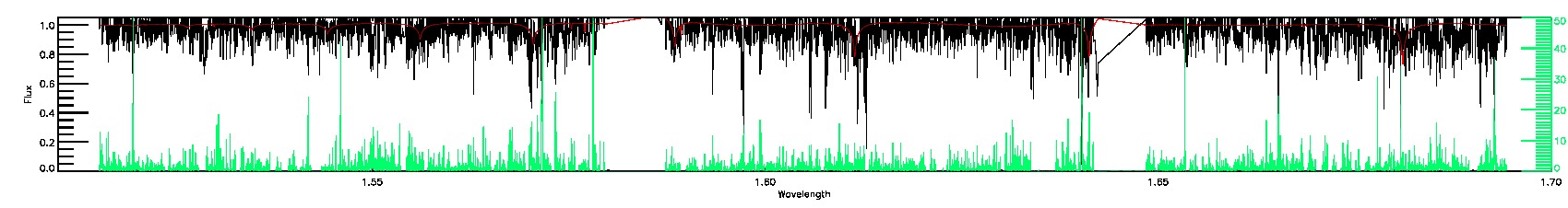
LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5756. | +/- | 94. |
| -10000. | +/- | -NaN |
| 2.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.02 | +/- | 1. |
| 1.02 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 18.67 | +/- | 0. |
| 1.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7324. | +/- | 128. |
| -10000. | +/- | -NaN |
| 3.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.66 | +/- | 0. |
| 4.66 | +/- | -NaN |
| -1.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 18.75 | +/- | 0. |
| 1.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.12 |
| -9999.00 |
LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5543. | +/- | 127. |
| -10000. | +/- | -NaN |
| 1.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| -1.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.37 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.66 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6082. | +/- | 100. |
| -10000. | +/- | -NaN |
| 3.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 21.71 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6093. | +/- | 51. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.76 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 3949. | +/- | 52. |
| -10000. | +/- | -NaN |
| 5.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -0.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 86.46 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6380. | +/- | 72. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.17 | +/- | 1. |
| 1.17 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 72.61 | +/- | 0. |
| 1.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.53 |
| -9999.00 |
BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6431. | +/- | 78. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.70 | +/- | 1. |
| 1.70 | +/- | -NaN |
| -1.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 62.88 | +/- | 0. |
| 1.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
BRIGHT_NEIGHBOR,VERY_BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 13725. | +/- | 1448. |
| -10000. | +/- | -NaN |
| 4.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.88 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.14 | +/- | 1. |
| 1.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.72 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.88 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 5288. | +/- | 61. |
| -10000. | +/- | -NaN |
| 1.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| -1.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.87 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.25 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.05 |
| -9999.00 |

BRIGHT_NEIGHBOR,VERY_BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 4265. | +/- | 47. |
| -10000. | +/- | -NaN |
| 0.97 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -1.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.64 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.66 |
| -9999.00 |

LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7474. | +/- | 165. |
| -10000. | +/- | -NaN |
| 3.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 85.21 | +/- | 0. |
| 1.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -2.29 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7134. | +/- | 125. |
| -10000. | +/- | -NaN |
| 3.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.36 | +/- | 0. |
| 4.36 | +/- | -NaN |
| -0.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 14.44 | +/- | 0. |
| 1.16 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6596. | +/- | 87. |
| -10000. | +/- | -NaN |
| 2.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.12 | +/- | 0. |
| 3.12 | +/- | -NaN |
| -1.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.48 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -1.53 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6603. | +/- | 101. |
| -10000. | +/- | -NaN |
| 2.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.22 | +/- | 1. |
| 1.22 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.53 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.29 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6486. | +/- | 107. |
| -10000. | +/- | -NaN |
| 2.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 62.62 | +/- | 0. |
| 1.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.45 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7998. | +/- | 274. |
| -10000. | +/- | -NaN |
| 3.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 1. |
| 0.98 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 94.06 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.02 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6533. | +/- | 92. |
| -10000. | +/- | -NaN |
| 2.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.64 | +/- | 0. |
| 3.64 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.68 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.26 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 4796. | +/- | 117. |
| -10000. | +/- | -NaN |
| 0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.01 | +/- | 1. |
| 1.01 | +/- | -NaN |
| -1.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.41 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.63 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.40 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -2.46 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 7322. | +/- | 63. |
| -10000. | +/- | -NaN |
| 3.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.79 | +/- | 0. |
| 4.79 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 27.78 | +/- | 0. |
| 1.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 1.44 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.58 |
| -9999.00 |
LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6126. | +/- | 89. |
| -10000. | +/- | -NaN |
| 2.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.76 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.18 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5962. | +/- | 86. |
| -10000. | +/- | -NaN |
| 2.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.53 | +/- | 1. |
| 4.53 | +/- | -NaN |
| -2.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 16. |
| -9999.99 | +/- | -NaN |
| 0.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.74 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.86 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.37 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5574. | +/- | 120. |
| -10000. | +/- | -NaN |
| 1.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.77 | +/- | 0. |
| 4.77 | +/- | -NaN |
| -1.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.93 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |

LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6024. | +/- | 53. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.29 | +/- | 0. |
| 2.29 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.17 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 1.41 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.14 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5735. | +/- | 125. |
| -10000. | +/- | -NaN |
| 2.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.21 | +/- | 6. |
| 1.21 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 12. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.51 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.48 |
| -9999.00 |
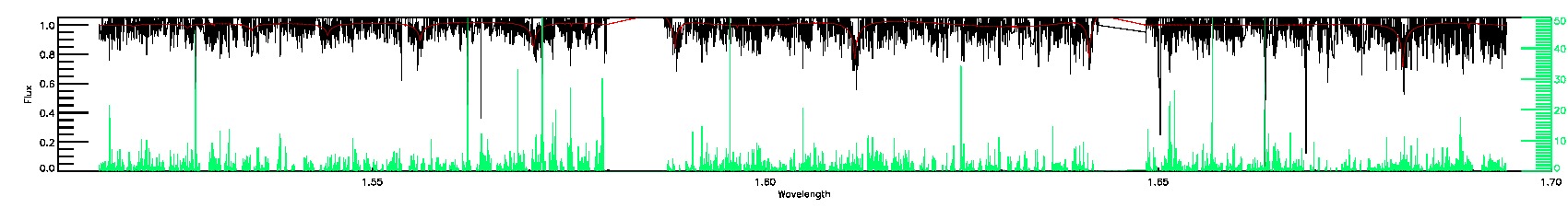
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 4442. | +/- | 24. |
| -10000. | +/- | -NaN |
| 2.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.06 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.53 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6437. | +/- | 110. |
| -10000. | +/- | -NaN |
| 3.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 12. |
| -9999.99 | +/- | -NaN |
| 0.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5341. | +/- | 113. |
| -10000. | +/- | -NaN |
| 1.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.81 | +/- | 3. |
| 0.81 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 9. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 12.54 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.45 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 1.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |

LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5949. | +/- | 127. |
| -10000. | +/- | -NaN |
| 2.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.65 | +/- | 1. |
| 0.65 | +/- | -NaN |
| -1.81 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.28 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.52 | +/- | 0. |
| 0.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.34 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5538. | +/- | 59. |
| -10000. | +/- | -NaN |
| 2.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 1. |
| 0.96 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 16.98 | +/- | 0. |
| 1.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.36 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5540. | +/- | 107. |
| -10000. | +/- | -NaN |
| 2.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.57 | +/- | 0. |
| 0.57 | +/- | -NaN |
| -1.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.68 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 19. |
| -9999.99 | +/- | -NaN |
| 0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.40 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.78 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.78 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6232. | +/- | 64. |
| -10000. | +/- | -NaN |
| 2.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.62 | +/- | 0. |
| 4.62 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.26 | +/- | 0. |
| 1.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 3770. | +/- | 21. |
| -10000. | +/- | -NaN |
| 0.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -1.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.42 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.92 |
| -9999.00 |
BRIGHT_NEIGHBOR,VERY_BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 8791. | +/- | 488. |
| -10000. | +/- | -NaN |
| 3.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 49.14 | +/- | 0. |
| 1.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.94 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.41 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 16358. | +/- | 1598. |
| -10000. | +/- | -NaN |
| 3.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.72 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 94.60 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.73 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.80 |
| -9999.00 |
BRIGHT_NEIGHBOR,LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7374. | +/- | 151. |
| -10000. | +/- | -NaN |
| 3.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.42 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.05 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 3898. | +/- | 81. |
| -10000. | +/- | -NaN |
| 4.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.56 | +/- | 1. |
| 1.56 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.96 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.53 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6441. | +/- | 45. |
| -10000. | +/- | -NaN |
| 2.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.67 | +/- | 1. |
| 1.67 | +/- | -NaN |
| -1.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.41 | +/- | 9. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 49.28 | +/- | 0. |
| 1.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.47 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6503. | +/- | 119. |
| -10000. | +/- | -NaN |
| 2.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.25 | +/- | 1. |
| 1.25 | +/- | -NaN |
| -1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 94.38 | +/- | 0. |
| 1.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.38 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5866. | +/- | 112. |
| -10000. | +/- | -NaN |
| 2.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.89 | +/- | 0. |
| 2.89 | +/- | -NaN |
| -1.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.69 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 1.07 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -2.47 |
| -9999.00 |
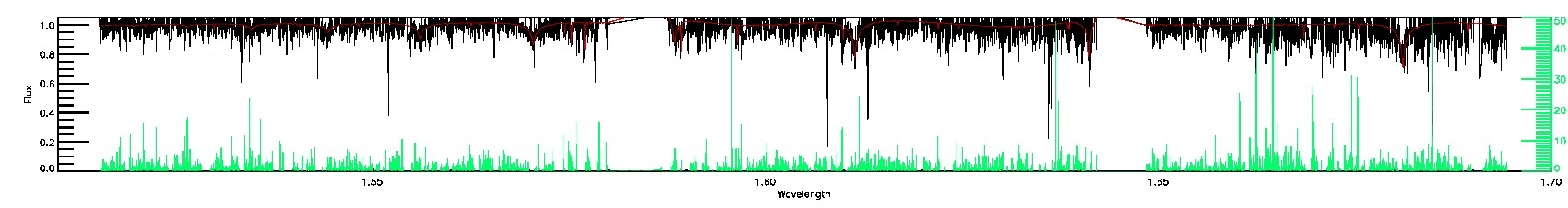
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6390. | +/- | 53. |
| -10000. | +/- | -NaN |
| 2.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| -0.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 28.89 | +/- | 0. |
| 1.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7929. | +/- | 154. |
| -10000. | +/- | -NaN |
| 3.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.86 | +/- | 0. |
| 1.86 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.99 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.97 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6333. | +/- | 41. |
| -10000. | +/- | -NaN |
| 2.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 2. |
| 1.49 | +/- | -NaN |
| -1.94 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 38.34 | +/- | 0. |
| 1.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6186. | +/- | 87. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -1.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.69 | +/- | 0. |
| 1.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.95 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6591. | +/- | 107. |
| -10000. | +/- | -NaN |
| 2.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 23.13 | +/- | 0. |
| 1.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.82 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5453. | +/- | 78. |
| -10000. | +/- | -NaN |
| 1.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.06 | +/- | 1. |
| 4.06 | +/- | -NaN |
| -1.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.31 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.05 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6424. | +/- | 84. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.58 | +/- | 1. |
| 0.58 | +/- | -NaN |
| -1.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.72 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.42 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7583. | +/- | 141. |
| -10000. | +/- | -NaN |
| 3.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.16 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.80 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6518. | +/- | 105. |
| -10000. | +/- | -NaN |
| 2.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.45 | +/- | 0. |
| 4.45 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 22.96 | +/- | 0. |
| 1.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.83 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5759. | +/- | 90. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.87 | +/- | 1. |
| 0.87 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 18.24 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.83 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6222. | +/- | 105. |
| -10000. | +/- | -NaN |
| 2.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 1. |
| 0.94 | +/- | -NaN |
| -0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 34.20 | +/- | 0. |
| 1.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6198. | +/- | 104. |
| -10000. | +/- | -NaN |
| 2.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.75 | +/- | 1. |
| 0.75 | +/- | -NaN |
| -1.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.09 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.83 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5787. | +/- | 90. |
| -10000. | +/- | -NaN |
| 1.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.21 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |

LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5086. | +/- | 40. |
| -10000. | +/- | -NaN |
| 1.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 | +/- | 1. |
| 0.43 | +/- | -NaN |
| -1.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.33 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.43 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6208. | +/- | 106. |
| -10000. | +/- | -NaN |
| 2.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.77 | +/- | 1. |
| 1.77 | +/- | -NaN |
| -1.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 1.64 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.98 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7995. | +/- | 163. |
| -10000. | +/- | -NaN |
| 3.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 10. |
| 0.83 | +/- | -NaN |
| -1.84 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 10.18 | +/- | 2. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.66 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7435. | +/- | 212. |
| -10000. | +/- | -NaN |
| 3.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 36.86 | +/- | 0. |
| 1.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| 0.75 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6653. | +/- | 80. |
| -10000. | +/- | -NaN |
| 2.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 1. |
| 0.52 | +/- | -NaN |
| -1.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.91 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.12 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.75 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5879. | +/- | 78. |
| -10000. | +/- | -NaN |
| 2.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| -1.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.18 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.74 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.77 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 7967. | +/- | 103. |
| -10000. | +/- | -NaN |
| 3.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.68 | +/- | 2. |
| 2.68 | +/- | -NaN |
| -1.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.42 | +/- | 2. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6302. | +/- | 57. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.93 | +/- | 0. |
| 1.93 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.96 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.31 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6645. | +/- | 98. |
| -10000. | +/- | -NaN |
| 3.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.05 | +/- | 1. |
| 1.05 | +/- | -NaN |
| -0.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.13 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.37 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 5968. | +/- | 70. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.64 | +/- | 0. |
| 2.64 | +/- | -NaN |
| -2.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.12 | +/- | 0. |
| 1.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.28 |
| -9999.00 |

LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7732. | +/- | 199. |
| -10000. | +/- | -NaN |
| 2.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.77 | +/- | 0. |
| 4.77 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 40.69 | +/- | 0. |
| 1.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.25 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7629. | +/- | 114. |
| -10000. | +/- | -NaN |
| 2.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.03 | +/- | 0. |
| 2.03 | +/- | -NaN |
| -1.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 10. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 25.51 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6751. | +/- | 118. |
| -10000. | +/- | -NaN |
| 2.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.54 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 30.88 | +/- | 0. |
| 1.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7759. | +/- | 155. |
| -10000. | +/- | -NaN |
| 3.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.95 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.52 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6263. | +/- | 69. |
| -10000. | +/- | -NaN |
| 2.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.74 | +/- | 0. |
| 3.74 | +/- | -NaN |
| -1.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 21.58 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.52 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6174. | +/- | 102. |
| -10000. | +/- | -NaN |
| 2.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 1. |
| 0.97 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 74.83 | +/- | 0. |
| 1.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5743. | +/- | 96. |
| -10000. | +/- | -NaN |
| 1.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.48 | +/- | 1. |
| 3.48 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 16. |
| -9999.99 | +/- | -NaN |
| 0.46 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 12.69 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.44 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5512. | +/- | 100. |
| -10000. | +/- | -NaN |
| 1.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.76 | +/- | 1. |
| 4.76 | +/- | -NaN |
| -2.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.22 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.56 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.50 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6337. | +/- | 117. |
| -10000. | +/- | -NaN |
| 3.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.66 | +/- | 1. |
| 0.66 | +/- | -NaN |
| -1.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 42.74 | +/- | 0. |
| 1.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6362. | +/- | 73. |
| -10000. | +/- | -NaN |
| 2.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.71 | +/- | 0. |
| 4.71 | +/- | -NaN |
| -0.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 14.38 | +/- | 0. |
| 1.16 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.42 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.58 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5650. | +/- | 107. |
| -10000. | +/- | -NaN |
| 1.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.68 | +/- | 1. |
| 0.68 | +/- | -NaN |
| -1.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.79 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.30 |
| -9999.00 |

LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7958. | +/- | 175. |
| -10000. | +/- | -NaN |
| 4.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.83 | +/- | 0. |
| 1.14 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5992. | +/- | 122. |
| -10000. | +/- | -NaN |
| 2.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.49 | +/- | 0. |
| 4.49 | +/- | -NaN |
| -1.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.96 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.39 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5840. | +/- | 71. |
| -10000. | +/- | -NaN |
| 2.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.12 | +/- | 0. |
| 1.12 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 22.97 | +/- | 0. |
| 1.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.82 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5724. | +/- | 129. |
| -10000. | +/- | -NaN |
| 3.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.44 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_JUMP_NEG,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5453. | +/- | 113. |
| -10000. | +/- | -NaN |
| 1.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 2. |
| 0.53 | +/- | -NaN |
| -2.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.82 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.08 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.33 |
| -9999.00 |

BRIGHT_NEIGHBOR,PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6434. | +/- | 125. |
| -10000. | +/- | -NaN |
| 3.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 1. |
| 0.76 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 91.37 | +/- | 0. |
| 1.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.39 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,LOW_SNR,PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 3804. | +/- | 0. |
| -10000. | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.71 | +/- | 0. |
| 4.71 | +/- | -NaN |
| 1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.65 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7487. | +/- | 105. |
| -10000. | +/- | -NaN |
| 3.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.56 | +/- | 4. |
| 1.56 | +/- | -NaN |
| -2.35 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 83. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 2.02 | +/- | 1. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.34 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6429. | +/- | 48. |
| -10000. | +/- | -NaN |
| 2.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.50 | +/- | 1. |
| 1.50 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 75.02 | +/- | 0. |
| 1.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5940. | +/- | 104. |
| -10000. | +/- | -NaN |
| 2.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.93 | +/- | 0. |
| 0.93 | +/- | -NaN |
| -1.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.58 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7420. | +/- | 95. |
| -10000. | +/- | -NaN |
| 3.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.41 | +/- | 1. |
| 0.41 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.54 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.58 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5949. | +/- | 96. |
| -10000. | +/- | -NaN |
| 2.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.90 | +/- | 1. |
| 3.90 | +/- | -NaN |
| -1.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.23 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.77 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.35 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6386. | +/- | 35. |
| -10000. | +/- | -NaN |
| 2.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.14 | +/- | 7. |
| 1.14 | +/- | -NaN |
| -2.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 36. |
| -9999.99 | +/- | -NaN |
| 0.37 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 94.86 | +/- | 1. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5856. | +/- | 163. |
| -10000. | +/- | -NaN |
| 2.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -1.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.64 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.61 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.57 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.49 |
| -9999.00 |

LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7479. | +/- | 163. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.39 | +/- | 1. |
| 1.39 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.35 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.79 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6126. | +/- | 110. |
| -10000. | +/- | -NaN |
| 3.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 1. |
| 0.76 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.96 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.48 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.41 |
| -9999.00 |
LOW_SNR,PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7994. | +/- | 223. |
| -10000. | +/- | -NaN |
| 3.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.77 | +/- | 1. |
| 0.77 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 20.28 | +/- | 0. |
| 1.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR,LOW_SNR,PERSIST_JUMP_POS,PERSIST_JUMP_NEG,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5884. | +/- | 164. |
| -10000. | +/- | -NaN |
| 3.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 1. |
| 0.97 | +/- | -NaN |
| -1.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 18.95 | +/- | 0. |
| 1.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.41 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5969. | +/- | 97. |
| -10000. | +/- | -NaN |
| 2.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.57 | +/- | 1. |
| 4.57 | +/- | -NaN |
| -1.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| 0.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.85 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.56 |
| -9999.00 |
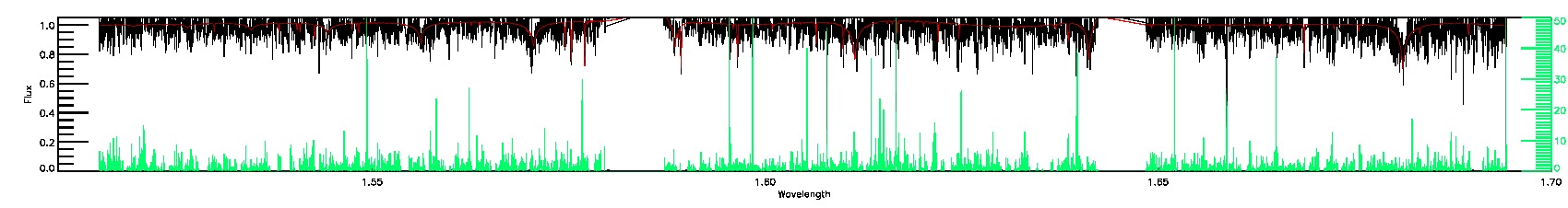
PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6633. | +/- | 124. |
| -10000. | +/- | -NaN |
| 2.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.96 | +/- | 1. |
| 1.96 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 42.31 | +/- | 0. |
| 1.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 4445. | +/- | 38. |
| -10000. | +/- | -NaN |
| 1.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.28 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.76 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5694. | +/- | 86. |
| -10000. | +/- | -NaN |
| 1.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 2. |
| 0.48 | +/- | -NaN |
| -2.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.72 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 14. |
| -9999.99 | +/- | -NaN |
| 0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.58 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.72 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.51 |
| -9999.00 |

BRIGHT_NEIGHBOR,LOW_SNR,PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6583. | +/- | 116. |
| -10000. | +/- | -NaN |
| 3.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.80 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
PERSIST_JUMP_NEG,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5811. | +/- | 99. |
| -10000. | +/- | -NaN |
| 2.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.79 | +/- | 3. |
| 4.79 | +/- | -NaN |
| -2.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 12. |
| -9999.99 | +/- | -NaN |
| 0.97 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 11.09 | +/- | 0. |
| 1.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.42 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5329. | +/- | 113. |
| -10000. | +/- | -NaN |
| 1.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.91 | +/- | 1. |
| 0.91 | +/- | -NaN |
| -2.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.57 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.31 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR,LOW_SNR,PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 4459. | +/- | 35. |
| -10000. | +/- | -NaN |
| 2.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.13 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.94 |
| -9999.00 |

STAR_WARN,SN_WARN
005-03-O
| 3758. | +/- | 7. |
| 3849. | +/- | 91. |
| 1.33 | +/- | 0. |
| 1.18 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.07 | +/- | 0. |
| 3.08 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.48 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7034. | +/- | 106. |
| -10000. | +/- | -NaN |
| 2.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.29 | +/- | 1. |
| 1.29 | +/- | -NaN |
| -0.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 43.48 | +/- | 0. |
| 1.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6340. | +/- | 101. |
| -10000. | +/- | -NaN |
| 2.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| -0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 22.04 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.10 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
005-03-O
| 4084. | +/- | 11. |
| -10000. | +/- | -NaN |
| 0.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.71 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.50 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7328. | +/- | 110. |
| -10000. | +/- | -NaN |
| 3.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.84 | +/- | 0. |
| 3.84 | +/- | -NaN |
| -1.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 21.53 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6075. | +/- | 125. |
| -10000. | +/- | -NaN |
| 2.79 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.56 | +/- | 0. |
| 4.56 | +/- | -NaN |
| -1.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 6. |
| -9999.99 | +/- | -NaN |
| 0.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 21.44 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.55 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5901. | +/- | 92. |
| -10000. | +/- | -NaN |
| 2.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 1. |
| 0.98 | +/- | -NaN |
| -1.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 45.26 | +/- | 0. |
| 1.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.61 |
| -9999.00 |
PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6270. | +/- | 108. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 1. |
| 0.98 | +/- | -NaN |
| -1.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 51.11 | +/- | 0. |
| 1.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.49 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7818. | +/- | 152. |
| -10000. | +/- | -NaN |
| 3.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.72 | +/- | 11. |
| 0.72 | +/- | -NaN |
| -2.19 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 3.33 | +/- | 2. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5969. | +/- | 105. |
| -10000. | +/- | -NaN |
| 2.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.22 | +/- | 7. |
| 1.22 | +/- | -NaN |
| -2.19 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.54 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 10.68 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.54 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -2.20 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6562. | +/- | 100. |
| -10000. | +/- | -NaN |
| 3.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.42 | +/- | 0. |
| 3.42 | +/- | -NaN |
| 0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.18 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.22 |
| -9999.00 |
PERSIST_JUMP_NEG,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5154. | +/- | 97. |
| -10000. | +/- | -NaN |
| 1.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.63 | +/- | 0. |
| 0.63 | +/- | -NaN |
| -1.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.99 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.87 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 2.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.38 |
| -9999.00 |

PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 4003. | +/- | 149. |
| -10000. | +/- | -NaN |
| 3.40 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 3.77 | +/- | 1. |
| 3.77 | +/- | -NaN |
| -2.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.02 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5534. | +/- | 132. |
| -10000. | +/- | -NaN |
| 2.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.37 | +/- | 0. |
| 3.37 | +/- | -NaN |
| -1.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.75 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.28 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.25 |
| -9999.00 |

STAR_WARN,SN_WARN
005-03-O
| 4025. | +/- | 5. |
| 4117. | +/- | 94. |
| 1.82 | +/- | 0. |
| 1.60 | +/- | 0. |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.09 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.13 |
| -9999.00 |

005-03-O
| 3788. | +/- | 1. |
| 3882. | +/- | 65. |
| 1.39 | +/- | 0. |
| 1.23 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.20 | +/- | 0. |
| 3.20 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.56 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6526. | +/- | 144. |
| -10000. | +/- | -NaN |
| 3.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.33 | +/- | 0. |
| 2.33 | +/- | -NaN |
| -1.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.77 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 28.00 | +/- | 0. |
| 1.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.11 |
| -9999.00 |
LOW_SNR,PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6537. | +/- | 127. |
| -10000. | +/- | -NaN |
| 5.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 1. |
| 0.76 | +/- | -NaN |
| 0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 91.66 | +/- | 0. |
| 1.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5915. | +/- | 119. |
| -10000. | +/- | -NaN |
| 3.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.80 | +/- | 0. |
| 2.80 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.19 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.90 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 1.29 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.21 |
| -9999.00 |

STAR_WARN,SN_WARN
005-03-O
| 3269. | +/- | 3. |
| 3374. | +/- | 77. |
| -0.23 | +/- | 0. |
| -0.32 | +/- | 0. |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.14 | +/- | 0. |
| 3.72 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.13 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6398. | +/- | 37. |
| -10000. | +/- | -NaN |
| 2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.66 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.74 | +/- | 0. |
| 1.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.15 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6162. | +/- | 152. |
| -10000. | +/- | -NaN |
| 2.99 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.68 | +/- | 1. |
| 0.68 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.85 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.77 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
LOW_SNR,PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7271. | +/- | 134. |
| -10000. | +/- | -NaN |
| 2.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 26.35 | +/- | 0. |
| 1.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.17 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 4946. | +/- | 136. |
| -10000. | +/- | -NaN |
| 2.90 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -1.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.71 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.62 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.47 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7998. | +/- | 111. |
| -10000. | +/- | -NaN |
| 3.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.80 | +/- | 3. |
| 2.80 | +/- | -NaN |
| -2.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 10. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 2.63 | +/- | 1. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.33 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5130. | +/- | 94. |
| -10000. | +/- | -NaN |
| 1.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.48 | +/- | 0. |
| 4.48 | +/- | -NaN |
| -1.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.26 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.32 | +/- | 0. |
| 0.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.52 |
| -9999.00 |

LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6727. | +/- | 128. |
| -10000. | +/- | -NaN |
| 2.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 85.27 | +/- | 0. |
| 1.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.42 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6535. | +/- | 93. |
| -10000. | +/- | -NaN |
| 2.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.62 | +/- | 1. |
| 3.62 | +/- | -NaN |
| -1.88 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.77 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 39.39 | +/- | 0. |
| 1.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5156. | +/- | 118. |
| -10000. | +/- | -NaN |
| 2.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 0. |
| 0.53 | +/- | -NaN |
| -1.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.48 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 1.50 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.69 |
| -9999.00 |

BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6996. | +/- | 113. |
| -10000. | +/- | -NaN |
| 3.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.46 | +/- | 0. |
| 3.46 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 71.65 | +/- | 0. |
| 1.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.48 |
| -9999.00 |
PERSIST_JUMP_NEG,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5745. | +/- | 86. |
| -10000. | +/- | -NaN |
| 1.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 | +/- | 1. |
| 1.00 | +/- | -NaN |
| -2.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 9.54 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -2.43 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6257. | +/- | 62. |
| -10000. | +/- | -NaN |
| 2.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.89 | +/- | 1. |
| 0.89 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.65 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5174. | +/- | 98. |
| -10000. | +/- | -NaN |
| 1.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -1.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.00 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.65 |
| -9999.00 |

LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5651. | +/- | 106. |
| -10000. | +/- | -NaN |
| 1.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.12 | +/- | 2. |
| 1.12 | +/- | -NaN |
| -2.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| -0.36 | +/- | 17. |
| -9999.99 | +/- | -NaN |
| 0.74 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 12.70 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -2.48 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5585. | +/- | 134. |
| -10000. | +/- | -NaN |
| 2.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.83 | +/- | 1. |
| 0.83 | +/- | -NaN |
| -1.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.62 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.26 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.65 |
| -9999.00 |

LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7926. | +/- | 162. |
| -10000. | +/- | -NaN |
| 3.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.95 | +/- | 0. |
| 1.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6410. | +/- | 68. |
| -10000. | +/- | -NaN |
| 2.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.72 | +/- | 0. |
| 0.72 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 60.23 | +/- | 0. |
| 1.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 1.00 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6230. | +/- | 110. |
| -10000. | +/- | -NaN |
| 3.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.08 | +/- | 0. |
| 3.08 | +/- | -NaN |
| 0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.86 | +/- | 0. |
| 1.14 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 1.32 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.99 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6961. | +/- | 89. |
| -10000. | +/- | -NaN |
| 2.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 48.03 | +/- | 0. |
| 1.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6023. | +/- | 92. |
| -10000. | +/- | -NaN |
| 2.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.79 | +/- | 0. |
| 4.79 | +/- | -NaN |
| -1.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.49 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.43 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5254. | +/- | 116. |
| -10000. | +/- | -NaN |
| 1.72 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.23 | +/- | 1. |
| 1.23 | +/- | -NaN |
| -1.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.69 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.66 |
| -9999.00 |

LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7640. | +/- | 191. |
| -10000. | +/- | -NaN |
| 3.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| 0.63 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 80.30 | +/- | 0. |
| 1.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7995. | +/- | 113. |
| -10000. | +/- | -NaN |
| 3.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.29 | +/- | 2. |
| 1.29 | +/- | -NaN |
| -1.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.15 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 10.44 | +/- | 1. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.88 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6617. | +/- | 108. |
| -10000. | +/- | -NaN |
| 3.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 1. |
| 0.33 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 28.95 | +/- | 0. |
| 1.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5353. | +/- | 101. |
| -10000. | +/- | -NaN |
| 1.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.90 | +/- | 1. |
| 0.90 | +/- | -NaN |
| -1.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 10. |
| -9999.99 | +/- | -NaN |
| 0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.89 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.56 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6890. | +/- | 85. |
| -10000. | +/- | -NaN |
| 2.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 44.44 | +/- | 0. |
| 1.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.41 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6492. | +/- | 91. |
| -10000. | +/- | -NaN |
| 2.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.12 | +/- | 0. |
| 4.12 | +/- | -NaN |
| -1.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 19.33 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.16 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5685. | +/- | 87. |
| -10000. | +/- | -NaN |
| 2.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.75 | +/- | 1. |
| 1.75 | +/- | -NaN |
| -1.92 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 49.37 | +/- | 0. |
| 1.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6275. | +/- | 139. |
| -10000. | +/- | -NaN |
| 3.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.93 | +/- | 5. |
| 1.93 | +/- | -NaN |
| -2.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 13. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 2.69 | +/- | 1. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.40 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6142. | +/- | 98. |
| -10000. | +/- | -NaN |
| 2.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.05 | +/- | 0. |
| 4.05 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 6. |
| -9999.99 | +/- | -NaN |
| 0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.67 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.85 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5881. | +/- | 85. |
| -10000. | +/- | -NaN |
| 2.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 83.77 | +/- | 0. |
| 1.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.01 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6514. | +/- | 99. |
| -10000. | +/- | -NaN |
| 2.71 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.82 | +/- | 2. |
| 0.82 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| -0.35 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 95.26 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.86 |
| -9999.00 |
LOW_SNR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 19784. | +/- | 1890. |
| -10000. | +/- | -NaN |
| 3.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -0.40 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.09 | +/- | 3. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.55 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5501. | +/- | 2. |
| -10000. | +/- | -NaN |
| 2.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.63 | +/- | 1. |
| 0.63 | +/- | -NaN |
| -1.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.91 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 1.19 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.58 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6321. | +/- | 87. |
| -10000. | +/- | -NaN |
| 2.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.89 | +/- | 1. |
| 0.89 | +/- | -NaN |
| -1.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.55 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.78 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6400. | +/- | 118. |
| -10000. | +/- | -NaN |
| 3.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.63 | +/- | 1. |
| 0.63 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 37.13 | +/- | 0. |
| 1.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.21 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5740. | +/- | 119. |
| -10000. | +/- | -NaN |
| 2.68 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 20.54 | +/- | 0. |
| 1.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.42 |
| -9999.00 |
BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5660. | +/- | 445. |
| -10000. | +/- | -NaN |
| 3.99 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.92 | +/- | 1. |
| 0.92 | +/- | -NaN |
| -0.94 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 23.17 | +/- | 0. |
| 1.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7016. | +/- | 129. |
| -10000. | +/- | -NaN |
| 2.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.66 | +/- | 1. |
| 2.66 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 5. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 90.09 | +/- | 0. |
| 1.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.29 |
| -9999.00 |
LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7226. | +/- | 137. |
| -10000. | +/- | -NaN |
| 3.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| 0.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 33.27 | +/- | 0. |
| 1.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.34 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_BAD,CHI2_BAD,ROTATION_BAD,SN_BAD
STAR_WARN,CHI2_WARN,ROTATION_WARN,SN_WARN
005-03-O
| 5426. | +/- | 101. |
| -10000. | +/- | -NaN |
| 1.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 4. |
| 0.52 | +/- | -NaN |
| -2.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.99 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| -0.38 | +/- | 7. |
| -9999.99 | +/- | -NaN |
| -0.39 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 12.38 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.35 |
| -9999.00 |

LOW_SNR
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7849. | +/- | 140. |
| -10000. | +/- | -NaN |
| 3.65 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.80 | +/- | 1. |
| 0.80 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.96 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 25.59 | +/- | 0. |
| 1.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.43 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5615. | +/- | 131. |
| -10000. | +/- | -NaN |
| 2.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| -0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 21.73 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 1.50 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7283. | +/- | 120. |
| -10000. | +/- | -NaN |
| 3.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.77 | +/- | 0. |
| 4.77 | +/- | -NaN |
| -1.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 4. |
| -9999.99 | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.27 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.76 |
| -9999.00 |
BRIGHT_NEIGHBOR,LOW_SNR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 5682. | +/- | 313. |
| -10000. | +/- | -NaN |
| 4.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.02 | +/- | 1. |
| 1.02 | +/- | -NaN |
| -1.54 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.65 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.42 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 4650. | +/- | 100. |
| -10000. | +/- | -NaN |
| 2.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -1.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.36 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.33 |
| -9999.00 |
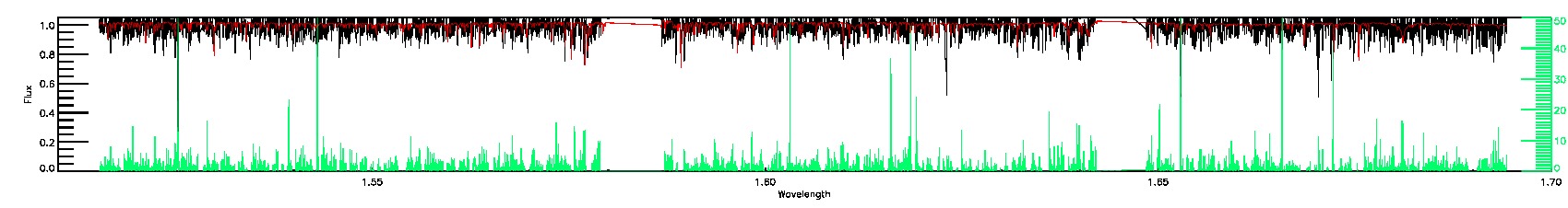
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7257. | +/- | 107. |
| -10000. | +/- | -NaN |
| 3.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.66 | +/- | 5. |
| 1.66 | +/- | -NaN |
| -2.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 6. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 3.47 | +/- | 2. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.46 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 6625. | +/- | 94. |
| -10000. | +/- | -NaN |
| 3.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.79 | +/- | 0. |
| 4.79 | +/- | -NaN |
| -1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.27 | +/- | 8. |
| -9999.99 | +/- | -NaN |
| 0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 15.41 | +/- | 0. |
| 1.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.51 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
005-03-O
| 7268. | +/- | 115. |
| -10000. | +/- | -NaN |
| 3.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.08 | +/- | 1. |
| 1.08 | +/- | -NaN |
| -1.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 12. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 26.05 | +/- | 0. |
| 1.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.64 |
| -9999.00 |
