STAR_WARN,SN_WARN
123-53-O
| 5132. | +/- | 20. |
| 5153. | +/- | 128. |
| 4.48 | +/- | 0. |
| 4.57 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 1.58 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.62 |
| -9999.00 |

123-53-O
| 5964. | +/- | 31. |
| 5890. | +/- | 153. |
| 4.12 | +/- | 0. |
| 4.07 | +/- | 0. |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 5.24 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.92 |
| -9999.00 |
123-53-O
| 6155. | +/- | 44. |
| 6054. | +/- | 170. |
| 4.40 | +/- | 0. |
| 4.26 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.08 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -2.38 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 5352. | +/- | 53. |
| 5368. | +/- | 149. |
| 4.34 | +/- | 0. |
| 4.58 | +/- | 0. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -0.61 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.18 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -0.59 |
| -9999.00 |

123-53-O
| 6006. | +/- | 20. |
| 5928. | +/- | 133. |
| 4.26 | +/- | 0. |
| 4.20 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.58 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 13.14 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.02 |
| -9999.00 |
123-53-O
| 5217. | +/- | 30. |
| 5242. | +/- | 136. |
| 3.52 | +/- | 0. |
| 3.44 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.90 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.20 |
| -9999.00 |

123-53-O
| 5757. | +/- | 26. |
| 5710. | +/- | 144. |
| 4.47 | +/- | 0. |
| 4.48 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.19 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.70 |
| -9999.00 |

123-53-O
| 6278. | +/- | 19. |
| 6163. | +/- | 145. |
| 4.28 | +/- | 0. |
| 4.12 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 7.96 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
123-53-O
| 6151. | +/- | 14. |
| 6052. | +/- | 130. |
| 4.28 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.50 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
123-53-O
| 5526. | +/- | 23. |
| 5506. | +/- | 134. |
| 4.20 | +/- | 0. |
| 4.25 | +/- | 0. |
| 0.82 | +/- | 0. |
| 0.82 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |

123-53-O
| 4418. | +/- | 6. |
| 4499. | +/- | 99. |
| 4.42 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.05 | +/- | 0. |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
123-53-O
| 5273. | +/- | 9. |
| 5259. | +/- | 112. |
| 4.44 | +/- | 0. |
| 4.32 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.30 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.50 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.41 |
| -9999.00 |

123-53-O
| 5733. | +/- | 24. |
| 5686. | +/- | 146. |
| 4.48 | +/- | 0. |
| 4.46 | +/- | 0. |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.12 | +/- | 0. |
| 1.54 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.36 |
| -9999.00 |

123-53-O
| 5534. | +/- | 20. |
| 5491. | +/- | 133. |
| 4.19 | +/- | 0. |
| 4.04 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.55 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
123-53-O
| 4226. | +/- | 5. |
| 4313. | +/- | 93. |
| 4.39 | +/- | 0. |
| 4.58 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.67 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |

SUSPECT_BROAD_LINES
123-53-O
| 6531. | +/- | 16. |
| 6386. | +/- | 145. |
| 4.33 | +/- | 0. |
| 4.12 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 12.20 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
123-53-O
| 4987. | +/- | 6. |
| 5022. | +/- | 92. |
| 3.51 | +/- | 0. |
| 3.37 | +/- | 0. |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.08 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 5835. | +/- | 42. |
| 5779. | +/- | 162. |
| 4.39 | +/- | 0. |
| 4.38 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.96 |
| -9999.00 |

123-53-O
| 5200. | +/- | 8. |
| 5206. | +/- | 98. |
| 4.43 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 6.55 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.14 |
| -9999.00 |

123-53-O
| 5939. | +/- | 18. |
| 5888. | +/- | 130. |
| 4.04 | +/- | 0. |
| 4.19 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -0.66 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.50 |
| -9999.00 |

123-53-O
| 4800. | +/- | 14. |
| 4887. | +/- | 117. |
| 4.34 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| -0.80 | +/- | 0. |
| -0.80 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.13 | +/- | 0. |
| 7.05 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.24 |
| -9999.00 |

123-53-O
| 5927. | +/- | 19. |
| 5864. | +/- | 136. |
| 4.30 | +/- | 0. |
| 4.31 | +/- | 0. |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.75 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5950. | +/- | 40. |
| 5880. | +/- | 166. |
| 4.42 | +/- | 0. |
| 4.38 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 1.76 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
123-53-O
| 5914. | +/- | 14. |
| 5839. | +/- | 129. |
| 4.58 | +/- | 0. |
| 4.47 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.69 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |

123-53-O
| 4217. | +/- | 3. |
| 4313. | +/- | 78. |
| 4.19 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 10.38 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |

123-53-O
| 5580. | +/- | 15. |
| 5543. | +/- | 126. |
| 4.39 | +/- | 0. |
| 4.33 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 1.85 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4594. | +/- | 10. |
| 4669. | +/- | 107. |
| 4.57 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.35 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 1.00 |
| -9999.00 |
123-53-O
| 5023. | +/- | 9. |
| 5045. | +/- | 105. |
| 4.54 | +/- | 0. |
| 4.53 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.65 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.34 |
| -9999.00 |
123-53-O
| 5049. | +/- | 8. |
| 5073. | +/- | 102. |
| 4.48 | +/- | 0. |
| 4.52 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.60 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.60 |
| -9999.00 |

123-53-O
| 4547. | +/- | 6. |
| 4632. | +/- | 99. |
| 4.50 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.72 | +/- | 0. |
| 0.72 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.74 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |

123-53-O
| 5846. | +/- | 32. |
| 5785. | +/- | 157. |
| 4.32 | +/- | 0. |
| 4.28 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.17 |
| -9999.00 |
BRIGHT_NEIGHBOR
123-53-O
| 5669. | +/- | 18. |
| 5610. | +/- | 137. |
| 4.42 | +/- | 0. |
| 4.24 | +/- | 0. |
| 0.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.80 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.37 |
| -9999.00 |

123-53-O
| 4733. | +/- | 8. |
| 4798. | +/- | 106. |
| 4.48 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.52 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
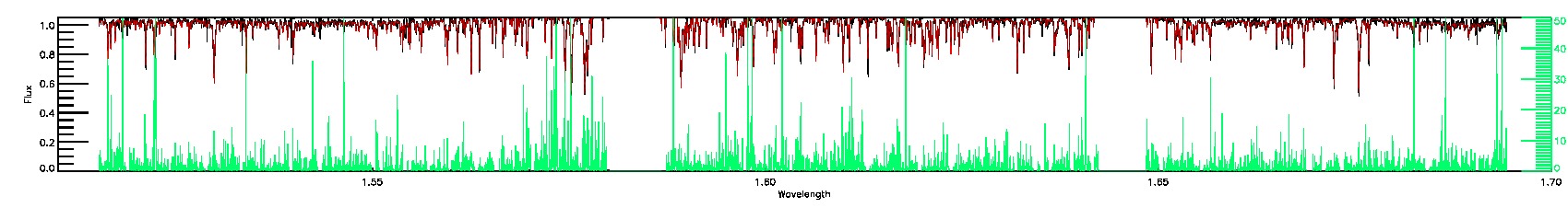
123-53-O
| 4514. | +/- | 8. |
| 4612. | +/- | 107. |
| 4.44 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.20 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.05 |
| -9999.00 |

123-53-O
| 5922. | +/- | 38. |
| 5856. | +/- | 162. |
| 4.37 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 6.11 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.52 |
| -9999.00 |
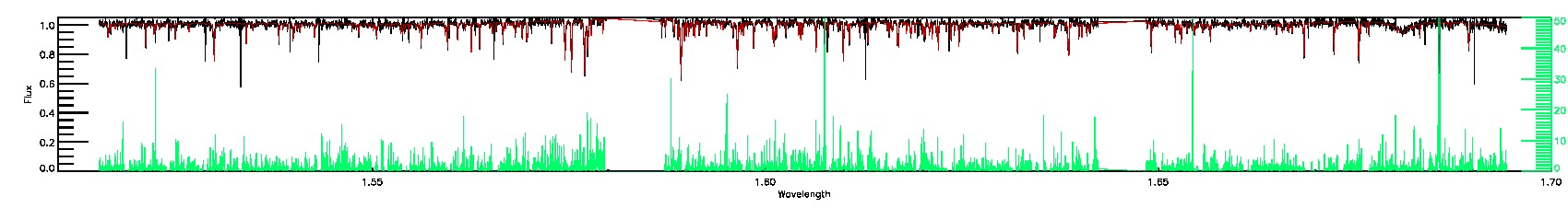
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
123-53-O
| 5477. | +/- | 89. |
| -10000. | +/- | -NaN |
| 4.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.89 | +/- | 0. |
| 0.89 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.25 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.39 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.42 |
| -9999.00 |

123-53-O
| 5784. | +/- | 7. |
| 5712. | +/- | 112. |
| 4.48 | +/- | 0. |
| 4.28 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.40 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5206. | +/- | 29. |
| 5229. | +/- | 135. |
| 3.68 | +/- | 0. |
| 3.68 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.68 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 1.00 |
| -9999.00 |

123-53-O
| 5795. | +/- | 18. |
| 5735. | +/- | 140. |
| 4.60 | +/- | 0. |
| 4.52 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 1.79 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 1.00 |
| -9999.00 |

123-53-O
| 4548. | +/- | 6. |
| 4649. | +/- | 103. |
| 2.67 | +/- | 0. |
| 2.50 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.21 | +/- | 0. |
| 3.82 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.55 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
123-53-O
| 6072. | +/- | 50. |
| 5991. | +/- | 164. |
| 4.63 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.58 | +/- | 0. |
| 0.58 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 13.49 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.02 |
| -9999.00 |
123-53-O
| 5601. | +/- | 31. |
| 5572. | +/- | 148. |
| 4.54 | +/- | 0. |
| 4.58 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 1.65 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -1.28 |
| -9999.00 |
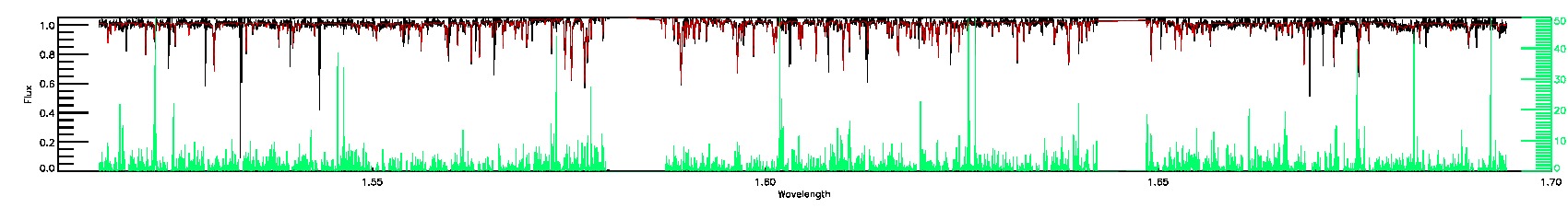
STAR_WARN,COLORTE_WARN
123-53-O
| 3366. | +/- | 3. |
| 3454. | +/- | 63. |
| 4.66 | +/- | 0. |
| 5.02 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.62 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 5.51 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.24 |
| -9999.00 |
123-53-O
| 4430. | +/- | 5. |
| 4527. | +/- | 98. |
| 4.28 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.22 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |

BRIGHT_NEIGHBOR
123-53-O
| 5630. | +/- | 19. |
| 5598. | +/- | 135. |
| 4.60 | +/- | 0. |
| 4.63 | +/- | 0. |
| 0.60 | +/- | 0. |
| 0.60 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.73 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.06 |
| -9999.00 |

123-53-O
| 6136. | +/- | 30. |
| 6038. | +/- | 157. |
| 4.27 | +/- | 0. |
| 4.15 | +/- | 0. |
| 0.48 | +/- | 0. |
| 0.48 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 10.63 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.41 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 4460. | +/- | 9. |
| 4549. | +/- | 107. |
| 4.40 | +/- | 0. |
| 4.57 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.22 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |

123-53-O
| 4649. | +/- | 4. |
| 4739. | +/- | 87. |
| 2.74 | +/- | 0. |
| 2.59 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.84 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5638. | +/- | 23. |
| 5582. | +/- | 142. |
| 4.51 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.70 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.28 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.99 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
123-53-O
| 3620. | +/- | 6. |
| 3718. | +/- | 83. |
| 4.51 | +/- | 0. |
| 4.91 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.54 |
| -9999.00 |
BRIGHT_NEIGHBOR
123-53-O
| 5845. | +/- | 36. |
| 5799. | +/- | 153. |
| 4.32 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.04 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.31 |
| -9999.00 |

123-53-O
| 5642. | +/- | 18. |
| 5598. | +/- | 136. |
| 4.42 | +/- | 0. |
| 4.35 | +/- | 0. |
| 1.23 | +/- | 0. |
| 1.23 | +/- | -NaN |
| -0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 1.54 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |

123-53-O
| 4295. | +/- | 2. |
| 4418. | +/- | 81. |
| 1.81 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.75 | +/- | 0. |
| 1.75 | +/- | -NaN |
| -0.82 | +/- | 0. |
| -0.82 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 4.77 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.78 |
| -9999.00 |
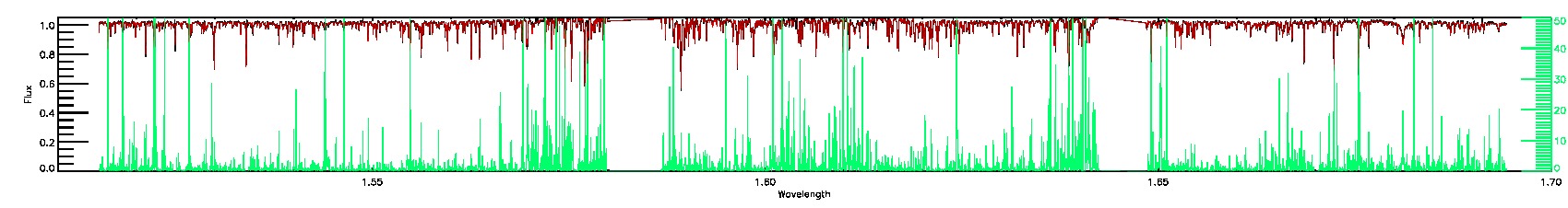
123-53-O
| 5880. | +/- | 20. |
| 5818. | +/- | 139. |
| 4.12 | +/- | 0. |
| 4.10 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.79 |
| -9999.00 |

BRIGHT_NEIGHBOR
123-53-O
| 5370. | +/- | 16. |
| 5361. | +/- | 124. |
| 4.68 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.98 | +/- | 0. |
| 0.98 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.21 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.95 |
| -9999.00 |

123-53-O
| 4309. | +/- | 2. |
| 4390. | +/- | 73. |
| 2.32 | +/- | 0. |
| 2.06 | +/- | 0. |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.65 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.30 |
| -9999.00 |

123-53-O
| 5590. | +/- | 14. |
| 5543. | +/- | 131. |
| 4.15 | +/- | 0. |
| 4.00 | +/- | 0. |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.03 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
123-53-O
| 6083. | +/- | 26. |
| 5985. | +/- | 153. |
| 4.57 | +/- | 0. |
| 4.39 | +/- | 0. |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.98 |
| -9999.00 |
123-53-O
| 5848. | +/- | 10. |
| 5771. | +/- | 124. |
| 4.44 | +/- | 0. |
| 4.23 | +/- | 0. |
| 0.74 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 5.74 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.53 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 5542. | +/- | 36. |
| 5509. | +/- | 147. |
| 4.48 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.58 |
| -9999.00 |
123-53-O
| 5177. | +/- | 22. |
| 5209. | +/- | 126. |
| 4.40 | +/- | 0. |
| 4.62 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.31 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.39 |
| -9999.00 |

123-53-O
| 5735. | +/- | 22. |
| 5683. | +/- | 142. |
| 4.55 | +/- | 0. |
| 4.49 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 1.83 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.86 |
| -9999.00 |

123-53-O
| 5757. | +/- | 31. |
| 5699. | +/- | 149. |
| 4.22 | +/- | 0. |
| 4.12 | +/- | 0. |
| 1.23 | +/- | 0. |
| 1.23 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.58 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.46 |
| -9999.00 |

123-53-O
| 5859. | +/- | 17. |
| 5787. | +/- | 135. |
| 4.22 | +/- | 0. |
| 4.09 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 13.48 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.20 |
| -9999.00 |

123-53-O
| 4170. | +/- | 2. |
| 4277. | +/- | 82. |
| 4.28 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.63 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.27 |
| -9999.00 |

123-53-O
| 6008. | +/- | 15. |
| 5923. | +/- | 127. |
| 4.42 | +/- | 0. |
| 4.31 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.20 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.29 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.08 |
| -9999.00 |
123-53-O
| 3874. | +/- | 5. |
| 3976. | +/- | 88. |
| 4.43 | +/- | 0. |
| 4.82 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.61 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.20 |
| -9999.00 |
123-53-O
| 5157. | +/- | 10. |
| 5154. | +/- | 113. |
| 4.28 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.65 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 1.67 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.15 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
123-53-O
| 8131. | +/- | 37. |
| 7925. | +/- | 259. |
| 4.13 | +/- | 0. |
| 3.66 | +/- | 0. |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 87.34 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.22 |
| -9999.00 |
123-53-O
| 3937. | +/- | 1. |
| 4070. | +/- | 75. |
| 0.93 | +/- | 0. |
| 0.92 | +/- | 0. |
| 2.19 | +/- | 0. |
| 2.19 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -1.06 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.25 | +/- | 0. |
| 5.49 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.44 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 4759. | +/- | 14. |
| 4812. | +/- | 113. |
| 4.58 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.87 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.05 | +/- | 0. |
| 2.57 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
BRIGHT_NEIGHBOR
123-53-O
| 4900. | +/- | 7. |
| 4951. | +/- | 99. |
| 3.57 | +/- | 0. |
| 3.49 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 3.36 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.46 |
| -9999.00 |

123-53-O
| 4645. | +/- | 7. |
| 4722. | +/- | 102. |
| 3.04 | +/- | 0. |
| 2.87 | +/- | 0. |
| 0.89 | +/- | 0. |
| 0.89 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.14 | +/- | 0. |
| 3.22 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.49 |
| -9999.00 |
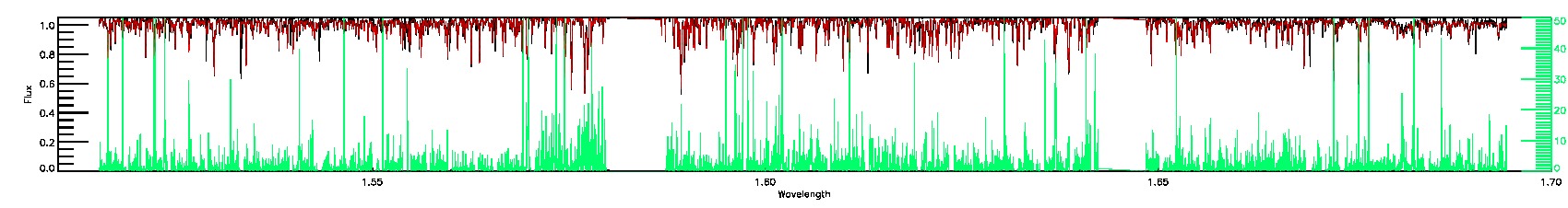
123-53-O
| 4955. | +/- | 12. |
| 5026. | +/- | 113. |
| 3.27 | +/- | 0. |
| 3.21 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.76 | +/- | -NaN |
| -0.84 | +/- | 0. |
| -0.84 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.28 | +/- | 0. |
| 4.82 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.65 |
| -9999.00 |

123-53-O
| 4727. | +/- | 5. |
| 4807. | +/- | 88. |
| 2.84 | +/- | 0. |
| 2.69 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -0.41 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.75 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.41 |
| -9999.00 |

123-53-O
| 4988. | +/- | 12. |
| 5036. | +/- | 109. |
| 4.26 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.60 | +/- | 0. |
| 0.60 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.37 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 6.06 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.39 |
| -9999.00 |

123-53-O
| 5856. | +/- | 18. |
| 5782. | +/- | 143. |
| 4.50 | +/- | 0. |
| 4.35 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.23 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.97 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.39 |
| -9999.00 |

BRIGHT_NEIGHBOR
123-53-O
| 6140. | +/- | 38. |
| 6057. | +/- | 176. |
| 4.74 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.17 | +/- | 0. |
| 1.59 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.58 |
| -9999.00 |
123-53-O
| 5645. | +/- | 16. |
| 5589. | +/- | 132. |
| 4.42 | +/- | 0. |
| 4.25 | +/- | 0. |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.29 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.59 |
| -9999.00 |

123-53-O
| 5292. | +/- | 20. |
| 5287. | +/- | 131. |
| 4.53 | +/- | 0. |
| 4.51 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.69 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
123-53-O
| 3632. | +/- | 2. |
| 3725. | +/- | 62. |
| 4.47 | +/- | 0. |
| 4.83 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 6.02 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
123-53-O
| 4343. | +/- | 4. |
| 4438. | +/- | 99. |
| 4.31 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.64 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |

123-53-O
| 5606. | +/- | 23. |
| 5571. | +/- | 131. |
| 4.35 | +/- | 0. |
| 4.33 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 12.03 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |

123-53-O
| 4922. | +/- | 6. |
| 4965. | +/- | 90. |
| 3.34 | +/- | 0. |
| 3.18 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.09 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.77 |
| -9999.00 |

123-53-O
| 5916. | +/- | 27. |
| 5862. | +/- | 143. |
| 4.09 | +/- | 0. |
| 4.18 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.20 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 3.99 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |

123-53-O
| 5160. | +/- | 15. |
| 5170. | +/- | 122. |
| 4.60 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.63 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.30 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.10 |
| -9999.00 |

123-53-O
| 5252. | +/- | 17. |
| 5256. | +/- | 128. |
| 4.51 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.52 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |

123-53-O
| 5966. | +/- | 22. |
| 5893. | +/- | 144. |
| 4.36 | +/- | 0. |
| 4.31 | +/- | 0. |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.20 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.99 |
| -9999.00 |
123-53-O
| 4586. | +/- | 7. |
| 4665. | +/- | 101. |
| 4.53 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.56 | +/- | 0. |
| 0.56 | +/- | -NaN |
| -0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.54 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.05 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5012. | +/- | 24. |
| 5066. | +/- | 131. |
| 3.50 | +/- | 0. |
| 3.43 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -0.60 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 4.19 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.19 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
123-53-O
| 3682. | +/- | 3. |
| 3783. | +/- | 68. |
| 4.49 | +/- | 0. |
| 4.91 | +/- | 0. |
| 0.74 | +/- | 0. |
| 0.74 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.21 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.35 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
123-53-O
| 3744. | +/- | 5. |
| 3844. | +/- | 82. |
| 4.50 | +/- | 0. |
| 4.91 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 1.00 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
123-53-O
| 7559. | +/- | 10. |
| 7366. | +/- | 199. |
| 4.19 | +/- | 0. |
| 3.85 | +/- | 0. |
| 4.36 | +/- | 0. |
| 4.36 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 14.68 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.42 |
| -9999.00 |
123-53-O
| 5341. | +/- | 26. |
| 5342. | +/- | 137. |
| 4.50 | +/- | 0. |
| 4.58 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.12 | +/- | 0. |
| 1.70 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.91 |
| -9999.00 |
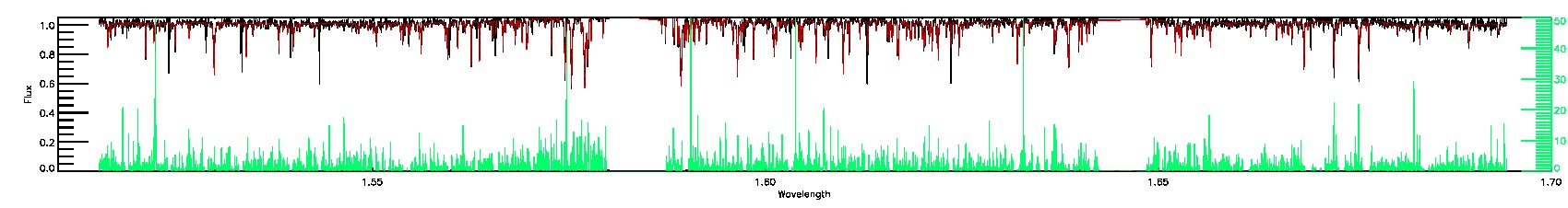
123-53-O
| 5038. | +/- | 18. |
| 5091. | +/- | 126. |
| 2.79 | +/- | 0. |
| 2.45 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -0.63 | +/- | 0. |
| -0.63 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.15 | +/- | 0. |
| 4.28 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.52 |
| -9999.00 |

123-53-O
| 5826. | +/- | 28. |
| 5763. | +/- | 154. |
| 4.25 | +/- | 0. |
| 4.17 | +/- | 0. |
| 1.34 | +/- | 0. |
| 1.34 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 1.77 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.68 |
| -9999.00 |
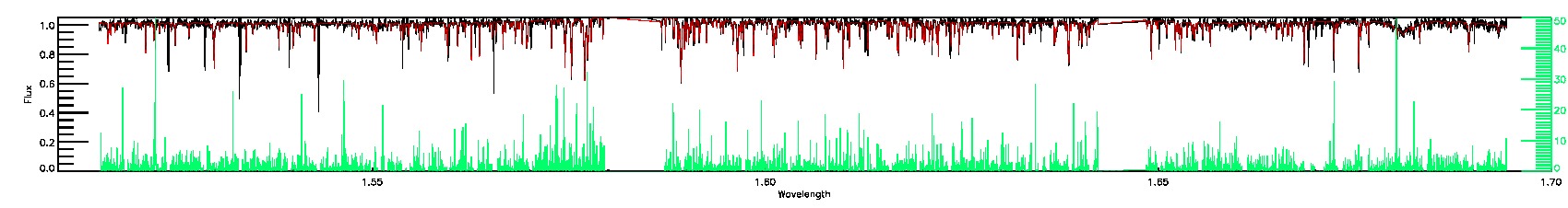
123-53-O
| 4426. | +/- | 2. |
| 4525. | +/- | 79. |
| 2.51 | +/- | 0. |
| 2.31 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.14 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.51 |
| -9999.00 |
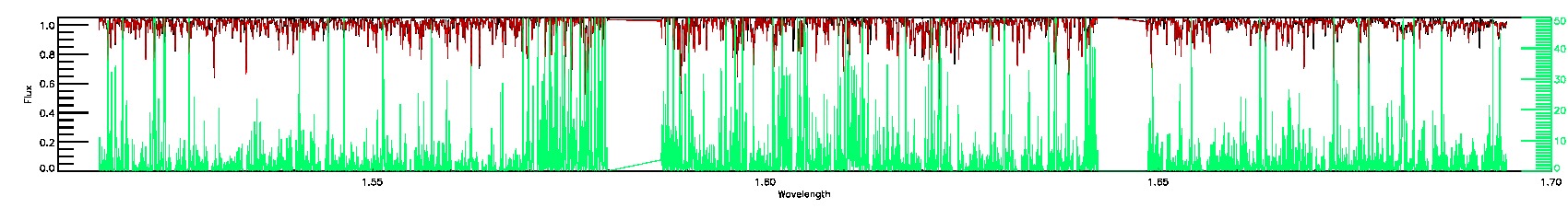
BRIGHT_NEIGHBOR
123-53-O
| 5126. | +/- | 9. |
| 5131. | +/- | 109. |
| 4.40 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 1.78 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.41 |
| -9999.00 |
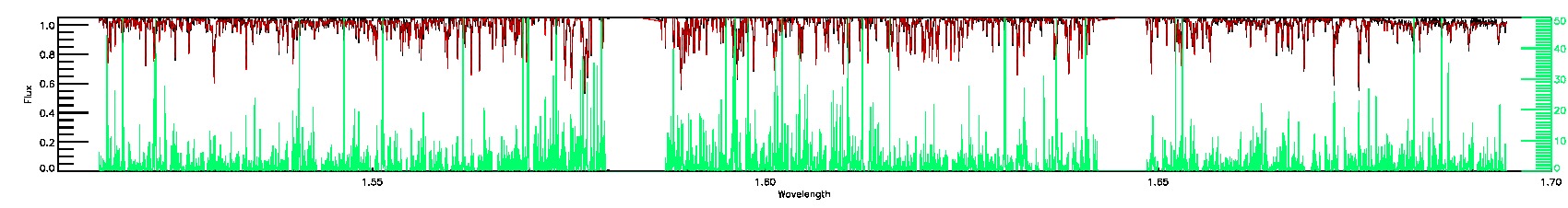
STAR_WARN,SN_WARN
123-53-O
| 3920. | +/- | 5. |
| 4019. | +/- | 91. |
| 4.40 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.10 | +/- | 0. |
| 2.66 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.50 |
| -9999.00 |
123-53-O
| 4408. | +/- | 6. |
| 4505. | +/- | 102. |
| 4.39 | +/- | 0. |
| 4.64 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.12 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.66 |
| -9999.00 |

123-53-O
| 5715. | +/- | 41. |
| 5681. | +/- | 153. |
| 4.21 | +/- | 0. |
| 4.30 | +/- | 0. |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 6.42 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.31 |
| -9999.00 |

123-53-O
| 4866. | +/- | 17. |
| 4923. | +/- | 120. |
| 3.20 | +/- | 0. |
| 3.05 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.41 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.15 |
| -9999.00 |

123-53-O
| 4515. | +/- | 7. |
| 4613. | +/- | 106. |
| 2.66 | +/- | 0. |
| 2.47 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 3.48 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.49 |
| -9999.00 |

123-53-O
| 4829. | +/- | 11. |
| 4905. | +/- | 107. |
| 4.17 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.68 | +/- | 0. |
| 0.68 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -0.61 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.89 |
| -9999.00 |

123-53-O
| 5850. | +/- | 22. |
| 5798. | +/- | 140. |
| 4.18 | +/- | 0. |
| 4.23 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.40 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.81 |
| -9999.00 |
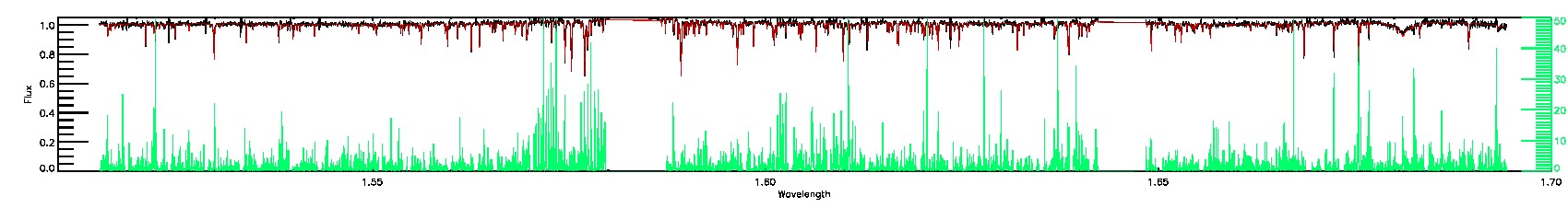
123-53-O
| 5007. | +/- | 7. |
| 5056. | +/- | 97. |
| 4.37 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.20 | +/- | 0. |
| 2.42 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.48 |
| -9999.00 |

123-53-O
| 5377. | +/- | 8. |
| 5358. | +/- | 107. |
| 4.63 | +/- | 0. |
| 4.55 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.14 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.18 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.55 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.14 |
| -9999.00 |

123-53-O
| 4708. | +/- | 4. |
| 4790. | +/- | 88. |
| 2.77 | +/- | 0. |
| 2.43 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 3.78 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.27 |
| -9999.00 |

123-53-O
| 5674. | +/- | 34. |
| 5643. | +/- | 153. |
| 3.86 | +/- | 0. |
| 3.90 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.73 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.53 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
123-53-O
| 6166. | +/- | 73. |
| -10000. | +/- | -NaN |
| 4.61 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.65 | +/- | 0. |
| 1.65 | +/- | -NaN |
| 0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.89 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.87 |
| -9999.00 |
STAR_WARN,COLORTE_WARN,SN_WARN
123-53-O
| 3820. | +/- | 6. |
| 3932. | +/- | 92. |
| 4.41 | +/- | 0. |
| 4.91 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -0.53 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.22 | +/- | 0. |
| 3.53 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.36 |
| -9999.00 |
123-53-O
| 3849. | +/- | 3. |
| 3936. | +/- | 74. |
| 4.51 | +/- | 0. |
| 4.76 | +/- | 0. |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.89 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.84 |
| -9999.00 |
123-53-O
| 6406. | +/- | 33. |
| 6282. | +/- | 179. |
| 4.81 | +/- | 0. |
| 4.67 | +/- | 0. |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 1.61 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.36 |
| -9999.00 |
123-53-O
| 5610. | +/- | 13. |
| 5570. | +/- | 118. |
| 4.51 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.61 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |

123-53-O
| 5009. | +/- | 8. |
| 5029. | +/- | 103. |
| 4.51 | +/- | 0. |
| 4.48 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 1.70 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.57 |
| -9999.00 |

SUSPECT_BROAD_LINES
123-53-O
| 6655. | +/- | 25. |
| 6510. | +/- | 167. |
| 4.34 | +/- | 0. |
| 4.24 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.38 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.19 | +/- | 0. |
| 39.34 | +/- | 0. |
| 1.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.26 |
| -9999.00 |
BRIGHT_NEIGHBOR
123-53-O
| 4673. | +/- | 5. |
| 4759. | +/- | 92. |
| 2.97 | +/- | 0. |
| 2.83 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -0.41 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.21 | +/- | 0. |
| 3.77 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.25 |
| -9999.00 |
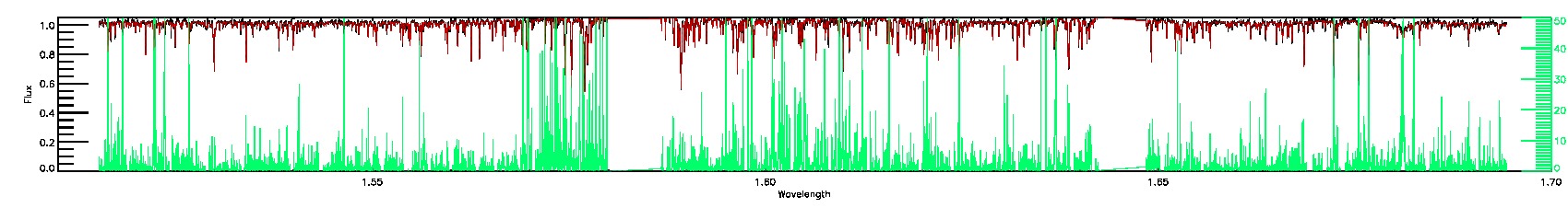
123-53-O
| 5024. | +/- | 13. |
| 5071. | +/- | 113. |
| 3.55 | +/- | 0. |
| 3.49 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.87 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.57 |
| -9999.00 |
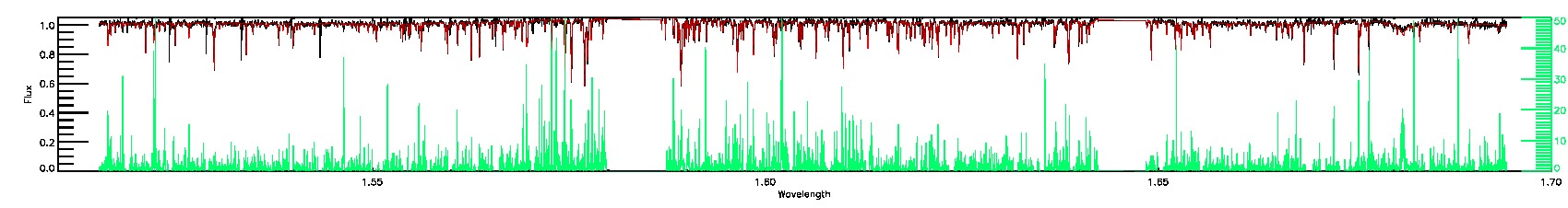
SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
123-53-O
| 3503. | +/- | 7. |
| 3594. | +/- | 78. |
| 4.59 | +/- | 0. |
| 4.95 | +/- | 0. |
| 1.73 | +/- | 0. |
| 1.73 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 31.82 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.95 |
| -9999.00 |
123-53-O
| 6670. | +/- | 23. |
| 6516. | +/- | 176. |
| 4.32 | +/- | 0. |
| 4.15 | +/- | 0. |
| 2.03 | +/- | 0. |
| 2.03 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 8.99 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.31 |
| -9999.00 |
123-53-O
| 5376. | +/- | 12. |
| 5378. | +/- | 110. |
| 4.53 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.00 | +/- | 0. |
| 1.67 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.25 |
| -9999.00 |

123-53-O
| 4731. | +/- | 5. |
| 4805. | +/- | 89. |
| 4.45 | +/- | 0. |
| 4.67 | +/- | 0. |
| 2.09 | +/- | 0. |
| 2.09 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 1.94 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.00 |
| -9999.00 |

123-53-O
| 4802. | +/- | 13. |
| 4852. | +/- | 113. |
| 4.29 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.70 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |

123-53-O
| 4437. | +/- | 6. |
| 4532. | +/- | 104. |
| 4.36 | +/- | 0. |
| 4.58 | +/- | 0. |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 3.19 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
STAR_WARN,COLORTE_WARN,SN_WARN
123-53-O
| 3811. | +/- | 6. |
| 3916. | +/- | 89. |
| 4.34 | +/- | 0. |
| 4.78 | +/- | 0. |
| 0.65 | +/- | 0. |
| 0.65 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.80 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -0.56 |
| -9999.00 |
123-53-O
| 5731. | +/- | 29. |
| 5672. | +/- | 148. |
| 4.43 | +/- | 0. |
| 4.30 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.73 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |

123-53-O
| 5956. | +/- | 15. |
| 5874. | +/- | 135. |
| 4.44 | +/- | 0. |
| 4.30 | +/- | 0. |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.48 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.02 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
123-53-O
| 3681. | +/- | 6. |
| 3791. | +/- | 87. |
| 4.29 | +/- | 0. |
| 4.81 | +/- | 0. |
| 0.58 | +/- | 0. |
| 0.58 | +/- | -NaN |
| -0.52 | +/- | 0. |
| -0.52 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.22 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.22 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
123-53-O
| 4452. | +/- | 7. |
| 4540. | +/- | 103. |
| 4.44 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.56 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |

123-53-O
| 5606. | +/- | 14. |
| 5570. | +/- | 124. |
| 4.43 | +/- | 0. |
| 4.41 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.19 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.71 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.37 |
| -9999.00 |

123-53-O
| 4547. | +/- | 5. |
| 4624. | +/- | 97. |
| 4.53 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.15 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.34 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.58 |
| -9999.00 |

123-53-O
| 6062. | +/- | 37. |
| 5973. | +/- | 162. |
| 4.48 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 8.81 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.76 |
| -9999.00 |
123-53-O
| 5888. | +/- | 38. |
| 5831. | +/- | 161. |
| 4.39 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -0.35 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.07 | +/- | 0. |
| 1.68 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.35 |
| -9999.00 |

123-53-O
| 5197. | +/- | 29. |
| 5218. | +/- | 133. |
| 4.22 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.57 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.12 |
| -9999.00 |
123-53-O
| 4536. | +/- | 6. |
| 4629. | +/- | 100. |
| 4.16 | +/- | 0. |
| 4.39 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.29 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.20 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 5555. | +/- | 41. |
| 5524. | +/- | 148. |
| 4.23 | +/- | 0. |
| 4.21 | +/- | 0. |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 6.06 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.10 |
| -9999.00 |

123-53-O
| 4685. | +/- | 3. |
| 4734. | +/- | 79. |
| 3.36 | +/- | 0. |
| 3.13 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.30 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.41 |
| -9999.00 |

123-53-O
| 5256. | +/- | 26. |
| 5271. | +/- | 134. |
| 4.50 | +/- | 0. |
| 4.63 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.23 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.09 |
| -9999.00 |

123-53-O
| 5490. | +/- | 24. |
| 5457. | +/- | 137. |
| 4.61 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 8.72 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.29 |
| -9999.00 |

123-53-O
| 4546. | +/- | 4. |
| 4648. | +/- | 84. |
| 4.37 | +/- | 0. |
| 4.68 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.77 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 5.16 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.36 |
| -9999.00 |

123-53-O
| 4382. | +/- | 4. |
| 4486. | +/- | 98. |
| 4.35 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.15 | +/- | 0. |
| 2.65 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.36 |
| -9999.00 |
123-53-O
| 6118. | +/- | 38. |
| 6022. | +/- | 164. |
| 4.63 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.57 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.98 |
| -9999.00 |
123-53-O
| 6125. | +/- | 17. |
| 6036. | +/- | 131. |
| 4.20 | +/- | 0. |
| 4.14 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 6.24 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.11 |
| -9999.00 |
123-53-O
| 6086. | +/- | 17. |
| 5988. | +/- | 135. |
| 4.30 | +/- | 0. |
| 4.13 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.51 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.04 |
| -9999.00 |
123-53-O
| 5713. | +/- | 26. |
| 5670. | +/- | 146. |
| 4.55 | +/- | 0. |
| 4.55 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.19 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.39 |
| -9999.00 |

123-53-O
| 4107. | +/- | 1. |
| 4223. | +/- | 77. |
| 1.75 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -0.66 | +/- | 0. |
| -0.66 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.23 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.61 |
| -9999.00 |

123-53-O
| 5396. | +/- | 9. |
| 5379. | +/- | 102. |
| 4.65 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.65 | +/- | 0. |
| 1.65 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.98 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.46 |
| -9999.00 |

123-53-O
| 4855. | +/- | 11. |
| 4897. | +/- | 109. |
| 4.59 | +/- | 0. |
| 4.62 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.57 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.43 |
| -9999.00 |

123-53-O
| 5532. | +/- | 18. |
| 5489. | +/- | 130. |
| 4.09 | +/- | 0. |
| 3.93 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.55 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.12 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
123-53-O
| 3782. | +/- | 6. |
| 3881. | +/- | 86. |
| 4.32 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.57 | +/- | 0. |
| 0.57 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 2.90 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.61 |
| -9999.00 |
123-53-O
| 4768. | +/- | 5. |
| 4845. | +/- | 90. |
| 2.90 | +/- | 0. |
| 2.76 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.19 | +/- | 0. |
| 3.88 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
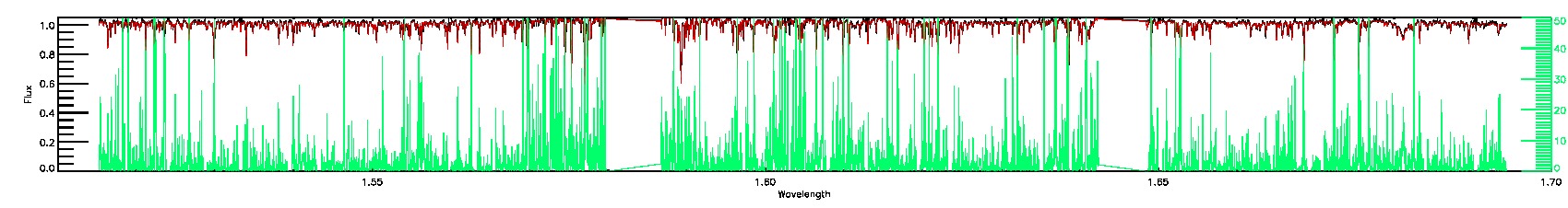
123-53-O
| 5474. | +/- | 11. |
| 5446. | +/- | 110. |
| 4.52 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.82 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 8.12 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.61 |
| -9999.00 |

123-53-O
| 5545. | +/- | 11. |
| 5509. | +/- | 117. |
| 4.53 | +/- | 0. |
| 4.45 | +/- | 0. |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.03 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.68 |
| -9999.00 |

123-53-O
| 5568. | +/- | 24. |
| 5532. | +/- | 141. |
| 4.52 | +/- | 0. |
| 4.46 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.83 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.33 |
| -9999.00 |

123-53-O
| 6510. | +/- | 27. |
| 6384. | +/- | 171. |
| 4.34 | +/- | 0. |
| 4.29 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.04 | +/- | 1. |
| 0.04 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.14 | +/- | 0. |
| 5.70 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.25 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
123-53-O
| 3739. | +/- | 5. |
| 3847. | +/- | 86. |
| 4.58 | +/- | 0. |
| 5.05 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.25 | +/- | 0. |
| 5.44 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 1.00 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 3896. | +/- | 6. |
| 3990. | +/- | 90. |
| 4.51 | +/- | 0. |
| 4.82 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 4788. | +/- | 15. |
| 4849. | +/- | 117. |
| 4.45 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.82 | +/- | 0. |
| 0.82 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.12 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 1.64 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.35 |
| -9999.00 |
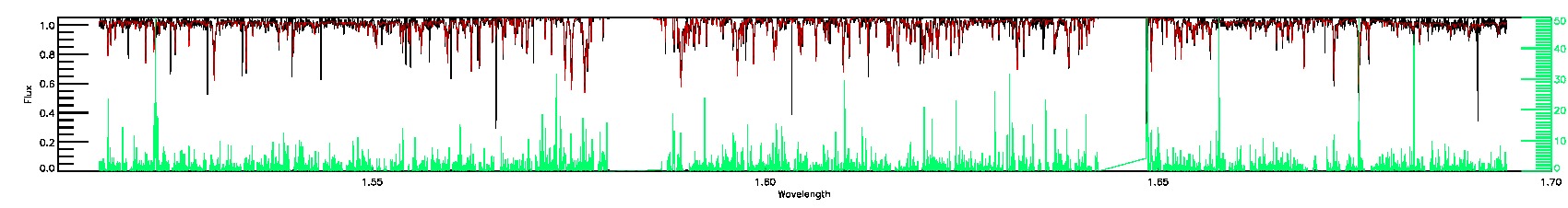
123-53-O
| 5889. | +/- | 20. |
| 5820. | +/- | 142. |
| 4.23 | +/- | 0. |
| 4.15 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.08 |
| -9999.00 |

123-53-O
| 5214. | +/- | 8. |
| 5217. | +/- | 102. |
| 4.48 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.85 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.13 |
| -9999.00 |

SUSPECT_BROAD_LINES
123-53-O
| 3458. | +/- | 4. |
| 3546. | +/- | 69. |
| 4.27 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 35.82 | +/- | 0. |
| 1.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.16 |
| -9999.00 |
123-53-O
| 5020. | +/- | 10. |
| 5047. | +/- | 106. |
| 3.65 | +/- | 0. |
| 3.56 | +/- | 0. |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.92 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.31 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4298. | +/- | 6. |
| 4400. | +/- | 103. |
| 4.23 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.27 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.36 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5280. | +/- | 41. |
| 5292. | +/- | 143. |
| 4.50 | +/- | 0. |
| 4.64 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 1.78 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.34 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5563. | +/- | 35. |
| 5535. | +/- | 146. |
| 4.56 | +/- | 0. |
| 4.57 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.49 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.99 |
| -9999.00 |
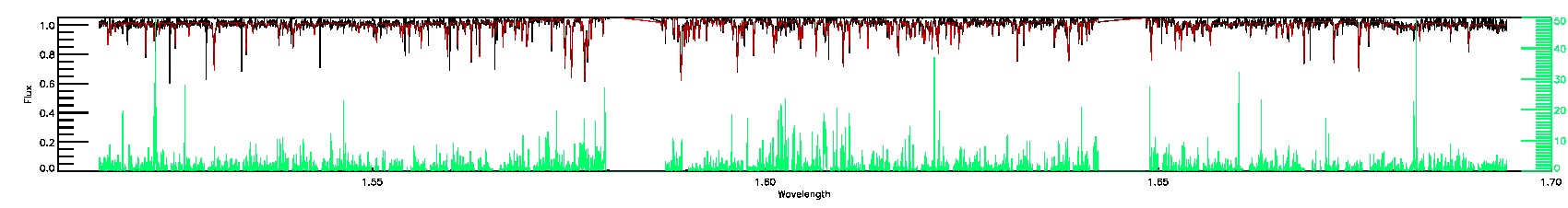
STAR_WARN,SN_WARN
123-53-O
| 5839. | +/- | 28. |
| 5769. | +/- | 154. |
| 4.60 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 2.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.40 |
| -9999.00 |

123-53-O
| 4110. | +/- | 1. |
| 4223. | +/- | 75. |
| 1.75 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.58 | +/- | -NaN |
| -0.57 | +/- | 0. |
| -0.57 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.22 | +/- | 0. |
| 4.14 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.71 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 6146. | +/- | 47. |
| 6049. | +/- | 174. |
| 4.55 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.63 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.78 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.76 |
| -9999.00 |
123-53-O
| 5758. | +/- | 13. |
| 5703. | +/- | 115. |
| 4.59 | +/- | 0. |
| 4.52 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.38 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.35 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 6000. | +/- | 47. |
| 5915. | +/- | 166. |
| 4.49 | +/- | 0. |
| 4.37 | +/- | 0. |
| 1.24 | +/- | 0. |
| 1.24 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.88 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.97 |
| -9999.00 |
123-53-O
| 5889. | +/- | 50. |
| 5842. | +/- | 166. |
| 4.47 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -0.61 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.18 | +/- | 0. |
| 1.61 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 1.00 |
| -9999.00 |

123-53-O
| 6006. | +/- | 36. |
| 5935. | +/- | 157. |
| 4.43 | +/- | 0. |
| 4.44 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.13 | +/- | 0. |
| 2.42 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.06 |
| -9999.00 |
SUSPECT_BROAD_LINES
123-53-O
| 5365. | +/- | 25. |
| 5349. | +/- | 132. |
| 4.42 | +/- | 0. |
| 4.37 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 16.24 | +/- | 0. |
| 1.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.32 |
| -9999.00 |
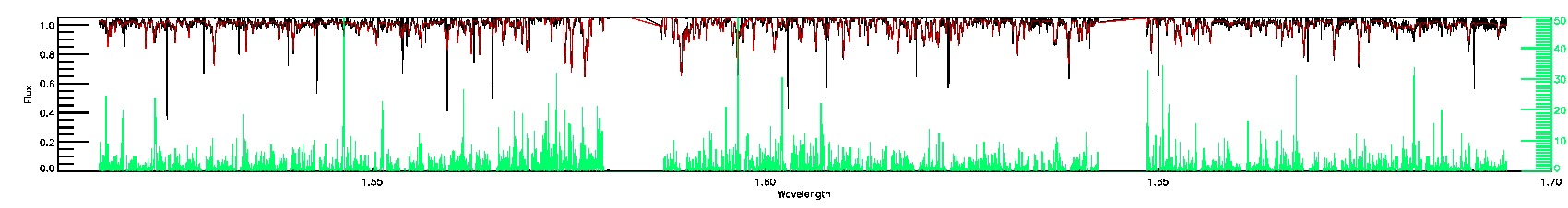
123-53-O
| 5642. | +/- | 24. |
| 5601. | +/- | 141. |
| 4.46 | +/- | 0. |
| 4.42 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 1.54 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.31 |
| -9999.00 |

123-53-O
| 5586. | +/- | 17. |
| 5559. | +/- | 129. |
| 4.54 | +/- | 0. |
| 4.58 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.14 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.03 | +/- | 0. |
| 1.51 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |

STAR_BAD
STAR_WARN,SN_WARN
123-53-O
| 5664. | +/- | 44. |
| -10000. | +/- | -NaN |
| 4.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.25 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |

STAR_BAD
STAR_WARN,COLORTE_WARN
123-53-O
| 3936. | +/- | 4. |
| -10000. | +/- | -NaN |
| 4.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.87 | +/- | 0. |
| 0.87 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.50 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
123-53-O
| 5864. | +/- | 22. |
| 5796. | +/- | 145. |
| 4.35 | +/- | 0. |
| 4.25 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.79 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.81 |
| -9999.00 |

123-53-O
| 5612. | +/- | 12. |
| 5582. | +/- | 113. |
| 4.05 | +/- | 0. |
| 4.09 | +/- | 0. |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.26 | +/- | 0. |
| 8.81 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
123-53-O
| 5788. | +/- | 11. |
| 5725. | +/- | 117. |
| 4.16 | +/- | 0. |
| 4.05 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 7.71 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.55 |
| -9999.00 |
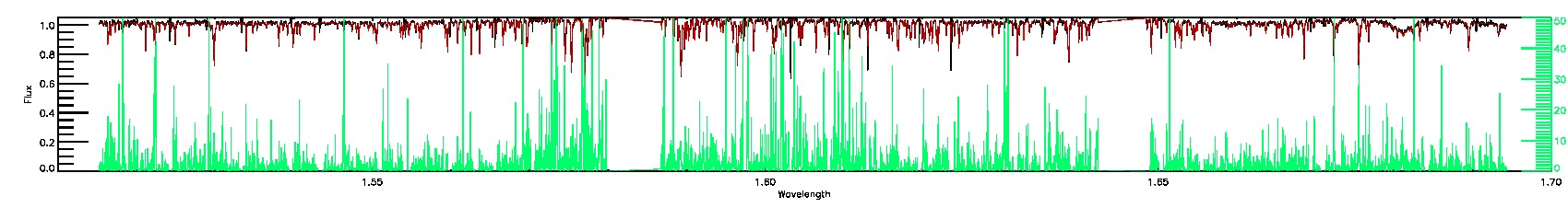
123-53-O
| 5536. | +/- | 15. |
| 5501. | +/- | 125. |
| 4.04 | +/- | 0. |
| 3.96 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.63 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.23 |
| -9999.00 |

123-53-O
| 5671. | +/- | 19. |
| 5619. | +/- | 131. |
| 4.34 | +/- | 0. |
| 4.22 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.74 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
123-53-O
| 3675. | +/- | 4. |
| 3781. | +/- | 80. |
| 4.44 | +/- | 0. |
| 4.90 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.38 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.99 |
| -9999.00 |
123-53-O
| 4529. | +/- | 7. |
| 4616. | +/- | 103. |
| 4.48 | +/- | 0. |
| 4.64 | +/- | 0. |
| 0.68 | +/- | 0. |
| 0.68 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.20 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
123-53-O
| 3640. | +/- | 4. |
| 3730. | +/- | 77. |
| 4.52 | +/- | 0. |
| 4.85 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 6.12 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.16 |
| -9999.00 |
123-53-O
| 4964. | +/- | 7. |
| 5024. | +/- | 97. |
| 2.81 | +/- | 0. |
| 2.47 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.76 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -0.59 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.22 | +/- | 0. |
| 4.18 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.30 |
| -9999.00 |

123-53-O
| 4745. | +/- | 5. |
| 4818. | +/- | 88. |
| 2.84 | +/- | 0. |
| 2.67 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.54 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.50 |
| -9999.00 |

123-53-O
| 5236. | +/- | 9. |
| 5235. | +/- | 107. |
| 4.53 | +/- | 0. |
| 4.49 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.60 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.86 |
| -9999.00 |

123-53-O
| 6426. | +/- | 18. |
| 6299. | +/- | 143. |
| 4.41 | +/- | 0. |
| 4.27 | +/- | 0. |
| 1.79 | +/- | 0. |
| 1.79 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.89 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.06 |
| -9999.00 |
123-53-O
| 3831. | +/- | 4. |
| 3925. | +/- | 82. |
| 4.37 | +/- | 0. |
| 4.70 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.93 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
123-53-O
| 3986. | +/- | 2. |
| 4077. | +/- | 70. |
| 4.41 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 8.30 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
123-53-O
| 6169. | +/- | 23. |
| 6065. | +/- | 156. |
| 4.71 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.70 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 2.37 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
SUSPECT_BROAD_LINES
123-53-O
| 4114. | +/- | 6. |
| 4194. | +/- | 83. |
| 4.34 | +/- | 0. |
| 4.48 | +/- | 0. |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.10 | +/- | 0. |
| 64.21 | +/- | 0. |
| 1.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |

123-53-O
| 5035. | +/- | 35. |
| 5129. | +/- | 143. |
| 2.71 | +/- | 0. |
| 2.71 | +/- | 0. |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| -1.60 | +/- | 0. |
| -1.60 | +/- | 0. |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.26 | +/- | 0. |
| 7.54 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.29 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4246. | +/- | 7. |
| 4335. | +/- | 98. |
| 4.36 | +/- | 0. |
| 4.57 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.40 | +/- | 0. |
| 7.23 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
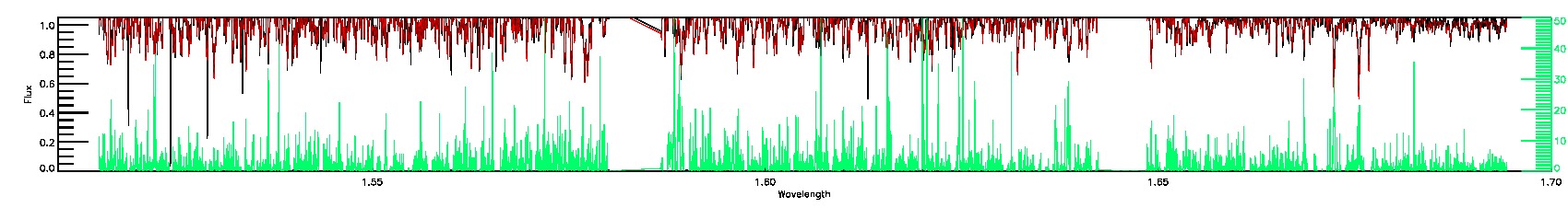
STAR_WARN,SN_WARN
123-53-O
| 5815. | +/- | 52. |
| 5768. | +/- | 165. |
| 4.12 | +/- | 0. |
| 4.18 | +/- | 0. |
| 0.89 | +/- | 0. |
| 0.89 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.99 |
| -9999.00 |

123-53-O
| 5470. | +/- | 19. |
| 5453. | +/- | 132. |
| 4.62 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 1.79 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.98 |
| -9999.00 |

STAR_BAD
123-53-O
| 4592. | +/- | 7. |
| -10000. | +/- | -NaN |
| 4.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.50 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |

123-53-O
| 5925. | +/- | 24. |
| 5848. | +/- | 150. |
| 4.38 | +/- | 0. |
| 4.26 | +/- | 0. |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.28 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |

123-53-O
| 6102. | +/- | 23. |
| 6001. | +/- | 148. |
| 4.41 | +/- | 0. |
| 4.22 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.25 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.78 |
| -9999.00 |
123-53-O
| 4822. | +/- | 6. |
| 4868. | +/- | 91. |
| 4.61 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.13 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.48 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.27 |
| -9999.00 |

123-53-O
| 5946. | +/- | 16. |
| 5867. | +/- | 137. |
| 4.13 | +/- | 0. |
| 4.01 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 4.84 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.26 |
| -9999.00 |

123-53-O
| 5664. | +/- | 12. |
| 5627. | +/- | 114. |
| 4.01 | +/- | 0. |
| 4.03 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.38 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.52 |
| -9999.00 |

123-53-O
| 4746. | +/- | 12. |
| 4813. | +/- | 112. |
| 4.55 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.25 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_WARN,SN_WARN
123-53-O
| 4561. | +/- | 13. |
| 4652. | +/- | 110. |
| 4.29 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 25.28 | +/- | 0. |
| 1.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5042. | +/- | 27. |
| 5063. | +/- | 128. |
| 4.65 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.70 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.12 |
| -9999.00 |

123-53-O
| 3840. | +/- | 2. |
| 3929. | +/- | 65. |
| 4.58 | +/- | 0. |
| 4.86 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.02 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.59 |
| -9999.00 |
123-53-O
| 5240. | +/- | 16. |
| 5238. | +/- | 125. |
| 4.62 | +/- | 0. |
| 4.58 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 1.65 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.85 |
| -9999.00 |

123-53-O
| 6142. | +/- | 16. |
| 6046. | +/- | 130. |
| 4.57 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.60 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.08 |
| -9999.00 |
123-53-O
| 5900. | +/- | 23. |
| 5839. | +/- | 137. |
| 4.46 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.73 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.93 |
| -9999.00 |
123-53-O
| 6252. | +/- | 18. |
| 6141. | +/- | 139. |
| 4.24 | +/- | 0. |
| 4.10 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.50 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 5.22 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.22 |
| -9999.00 |
123-53-O
| 5631. | +/- | 15. |
| 5593. | +/- | 121. |
| 4.42 | +/- | 0. |
| 4.39 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.80 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.48 |
| -9999.00 |

123-53-O
| 5962. | +/- | 17. |
| 5882. | +/- | 140. |
| 4.49 | +/- | 0. |
| 4.37 | +/- | 0. |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.91 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.84 |
| -9999.00 |
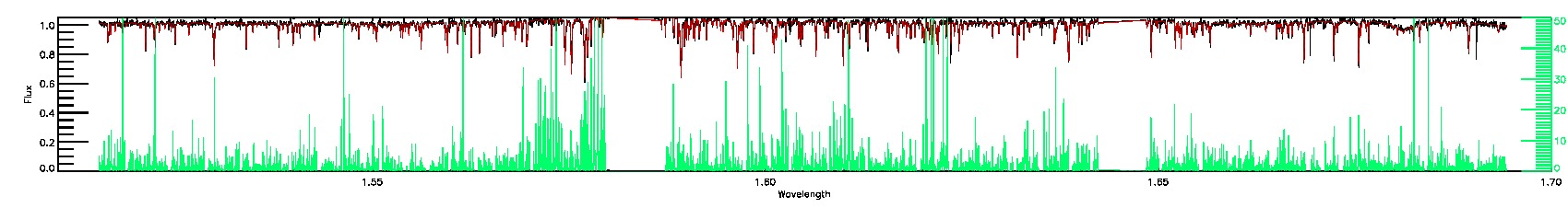
123-53-O
| 5302. | +/- | 13. |
| 5295. | +/- | 115. |
| 3.96 | +/- | 0. |
| 3.92 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.85 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |

123-53-O
| 4119. | +/- | 3. |
| 4210. | +/- | 80. |
| 4.36 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 6.20 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
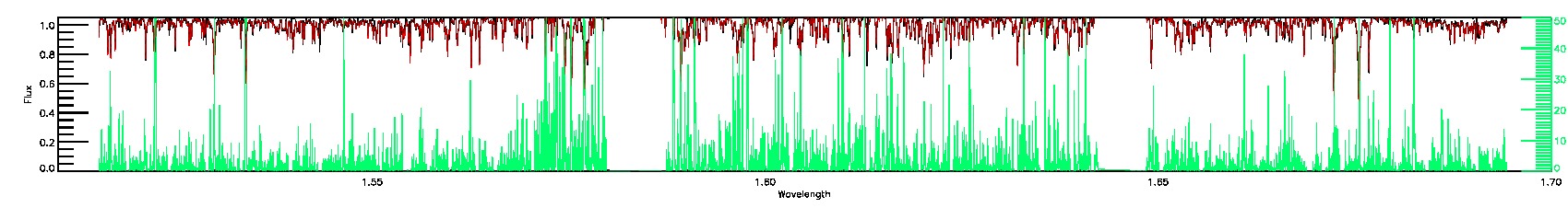
STAR_WARN,SN_WARN
123-53-O
| 4790. | +/- | 15. |
| 4849. | +/- | 117. |
| 4.38 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 2.17 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |

123-53-O
| 5024. | +/- | 15. |
| 5064. | +/- | 117. |
| 3.28 | +/- | 0. |
| 3.14 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.51 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.01 |
| -9999.00 |

123-53-O
| 6495. | +/- | 22. |
| 6372. | +/- | 160. |
| 4.29 | +/- | 0. |
| 4.25 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| 0.12 | +/- | 1. |
| 0.12 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.13 | +/- | 0. |
| 7.27 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.99 |
| -9999.00 |
123-53-O
| 6349. | +/- | 18. |
| 6230. | +/- | 139. |
| 4.54 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.65 | +/- | 0. |
| 0.65 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 8.37 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.31 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 4611. | +/- | 10. |
| 4681. | +/- | 107. |
| 4.54 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.90 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |

123-53-O
| 5188. | +/- | 26. |
| 5218. | +/- | 135. |
| 3.74 | +/- | 0. |
| 3.81 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.18 | +/- | 0. |
| 3.97 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.58 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4721. | +/- | 13. |
| 4776. | +/- | 112. |
| 4.58 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.77 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.06 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4642. | +/- | 11. |
| 4713. | +/- | 109. |
| 4.50 | +/- | 0. |
| 4.60 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.91 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.25 |
| -9999.00 |

123-53-O
| 4395. | +/- | 4. |
| 4491. | +/- | 94. |
| 4.34 | +/- | 0. |
| 4.57 | +/- | 0. |
| 0.59 | +/- | 0. |
| 0.59 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 6.66 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.10 |
| -9999.00 |
123-53-O
| 5891. | +/- | 26. |
| 5818. | +/- | 152. |
| 4.55 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.93 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4461. | +/- | 7. |
| 4541. | +/- | 101. |
| 4.54 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.07 | +/- | 0. |
| 1.82 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.06 |
| -9999.00 |

123-53-O
| 5956. | +/- | 10. |
| 5868. | +/- | 119. |
| 4.39 | +/- | 0. |
| 4.19 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.51 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.72 |
| -9999.00 |

123-53-O
| 4902. | +/- | 8. |
| 4933. | +/- | 99. |
| 3.72 | +/- | 0. |
| 3.60 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.53 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.01 |
| -9999.00 |
SUSPECT_BROAD_LINES
123-53-O
| 5609. | +/- | 25. |
| 5569. | +/- | 129. |
| 4.07 | +/- | 0. |
| 4.01 | +/- | 0. |
| 3.09 | +/- | 0. |
| 3.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 59.20 | +/- | 0. |
| 1.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.41 |
| -9999.00 |
123-53-O
| 4608. | +/- | 3. |
| 4681. | +/- | 81. |
| 2.69 | +/- | 0. |
| 2.38 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.85 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.01 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5194. | +/- | 34. |
| 5220. | +/- | 139. |
| 3.82 | +/- | 0. |
| 3.91 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.09 | +/- | 0. |
| 1.63 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -2.39 |
| -9999.00 |
123-53-O
| 4500. | +/- | 2. |
| 4597. | +/- | 81. |
| 2.77 | +/- | 0. |
| 2.59 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.20 | +/- | 0. |
| 3.35 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.24 |
| -9999.00 |
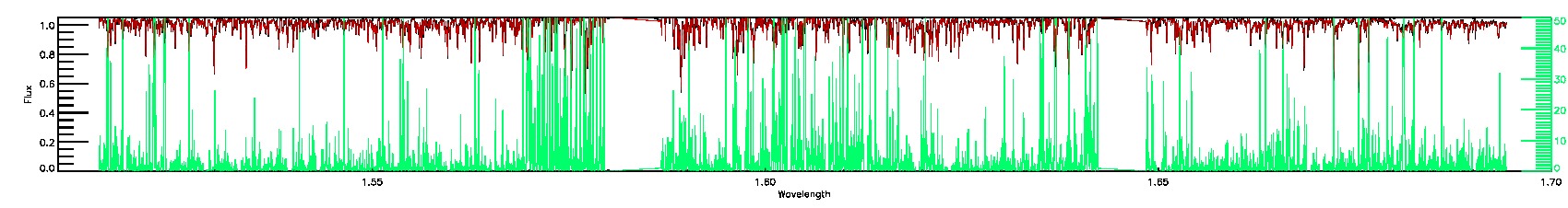
123-53-O
| 5756. | +/- | 33. |
| 5707. | +/- | 151. |
| 4.45 | +/- | 0. |
| 4.44 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 3.78 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.47 |
| -9999.00 |

123-53-O
| 5745. | +/- | 21. |
| 5687. | +/- | 138. |
| 4.13 | +/- | 0. |
| 4.02 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.26 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.41 |
| -9999.00 |

123-53-O
| 5880. | +/- | 17. |
| 5811. | +/- | 138. |
| 4.42 | +/- | 0. |
| 4.34 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.35 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.96 |
| -9999.00 |

123-53-O
| 6206. | +/- | 44. |
| 6106. | +/- | 174. |
| 4.35 | +/- | 0. |
| 4.28 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 6.43 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.06 |
| -9999.00 |
123-53-O
| 5836. | +/- | 37. |
| 5785. | +/- | 157. |
| 4.45 | +/- | 0. |
| 4.50 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.37 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.08 |
| -9999.00 |

123-53-O
| 5434. | +/- | 19. |
| 5433. | +/- | 123. |
| 4.46 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.86 | +/- | 0. |
| 0.86 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 2.54 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.27 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4529. | +/- | 8. |
| 4615. | +/- | 107. |
| 4.50 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.62 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.03 |
| -9999.00 |
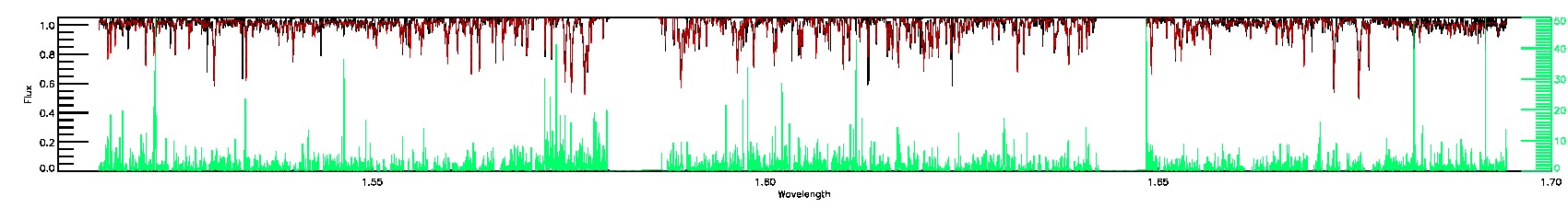
BRIGHT_NEIGHBOR
123-53-O
| 4361. | +/- | 5. |
| 4460. | +/- | 101. |
| 4.20 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 8.73 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.24 |
| -9999.00 |
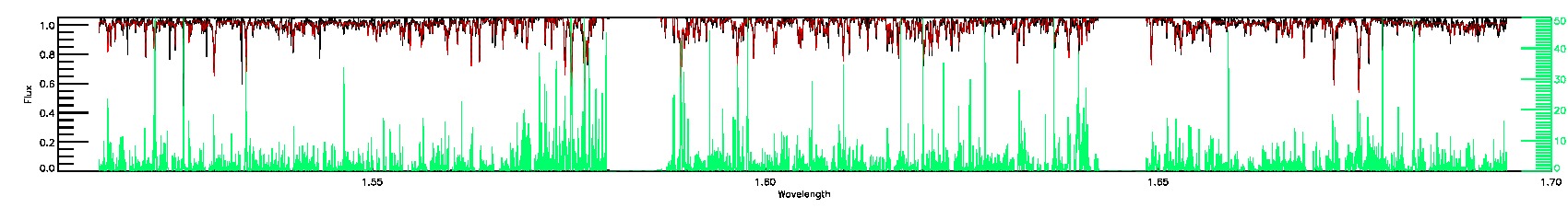
STAR_WARN,COLORTE_WARN,SN_WARN
123-53-O
| 3564. | +/- | 8. |
| 3660. | +/- | 84. |
| 4.47 | +/- | 0. |
| 4.87 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.00 | +/- | 0. |
| 4.22 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 5441. | +/- | 26. |
| 5420. | +/- | 138. |
| 4.39 | +/- | 0. |
| 4.36 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.83 |
| -9999.00 |

123-53-O
| 5198. | +/- | 26. |
| 5232. | +/- | 131. |
| 4.14 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| -0.63 | +/- | 0. |
| -0.62 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.21 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.45 |
| -9999.00 |
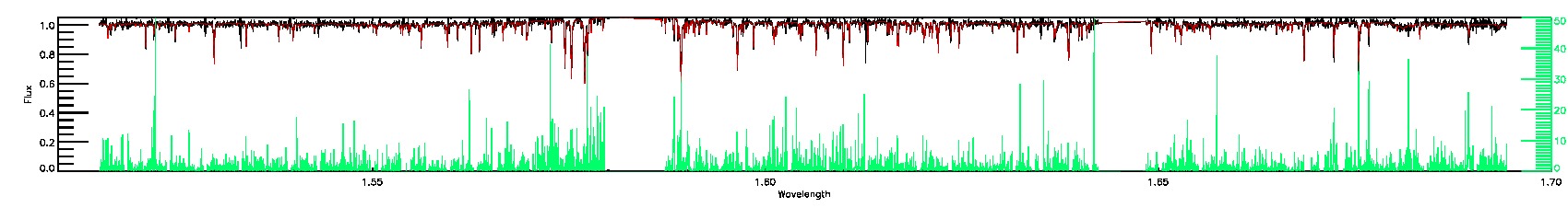
STAR_WARN,SN_WARN
123-53-O
| 5253. | +/- | 29. |
| 5266. | +/- | 136. |
| 4.40 | +/- | 0. |
| 4.53 | +/- | 0. |
| 0.89 | +/- | 0. |
| 0.89 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.21 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 5559. | +/- | 31. |
| 5523. | +/- | 144. |
| 4.41 | +/- | 0. |
| 4.34 | +/- | 0. |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.35 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.03 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 4669. | +/- | 14. |
| 4745. | +/- | 117. |
| 4.46 | +/- | 0. |
| 4.64 | +/- | 0. |
| 0.72 | +/- | 0. |
| 0.72 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.12 | +/- | 0. |
| 1.69 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
123-53-O
| 3456. | +/- | 10. |
| 3557. | +/- | 84. |
| 4.55 | +/- | 0. |
| 5.01 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.51 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 6.20 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.27 |
| -9999.00 |
123-53-O
| 6252. | +/- | 23. |
| 6143. | +/- | 146. |
| 4.86 | +/- | 0. |
| 4.74 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.13 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 10.24 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.26 |
| -9999.00 |
123-53-O
| 5734. | +/- | 36. |
| 5692. | +/- | 152. |
| 4.35 | +/- | 0. |
| 4.39 | +/- | 0. |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 2.98 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.90 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 4945. | +/- | 19. |
| 4994. | +/- | 124. |
| 4.31 | +/- | 0. |
| 4.48 | +/- | 0. |
| 0.61 | +/- | 0. |
| 0.61 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.12 | +/- | 0. |
| 2.14 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.17 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 5222. | +/- | 22. |
| 5226. | +/- | 132. |
| 4.50 | +/- | 0. |
| 4.51 | +/- | 0. |
| 1.13 | +/- | 0. |
| 1.13 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 1.63 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.03 |
| -9999.00 |

123-53-O
| 5500. | +/- | 12. |
| 5470. | +/- | 119. |
| 4.61 | +/- | 0. |
| 4.55 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 1.82 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.16 |
| -9999.00 |
123-53-O
| 4701. | +/- | 4. |
| 4766. | +/- | 83. |
| 4.57 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.07 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.21 |
| -9999.00 |

STAR_WARN,SN_WARN
123-53-O
| 5958. | +/- | 41. |
| 5891. | +/- | 168. |
| 4.38 | +/- | 0. |
| 4.38 | +/- | 0. |
| 1.86 | +/- | 0. |
| 1.86 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.31 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.14 | +/- | 0. |
| 1.83 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.86 |
| -9999.00 |
123-53-O
| 4206. | +/- | 5. |
| 4314. | +/- | 96. |
| 4.29 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.16 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.59 |
| -9999.00 |
STAR_WARN,COLORTE_WARN,SN_WARN
123-53-O
| 6253. | +/- | 44. |
| 6132. | +/- | 174. |
| 4.64 | +/- | 0. |
| 4.40 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 4.77 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.20 |
| -9999.00 |
STAR_WARN,SN_WARN
123-53-O
| 3736. | +/- | 11. |
| 3825. | +/- | 90. |
| 4.39 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 4.95 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
123-53-O
| 5799. | +/- | 45. |
| 5752. | +/- | 162. |
| 4.50 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.15 | +/- | 0. |
| 2.29 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.33 |
| -9999.00 |

123-53-O
| 6056. | +/- | 29. |
| 5967. | +/- | 155. |
| 4.40 | +/- | 0. |
| 4.28 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.80 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
123-53-O
| 5497. | +/- | 106. |
| -10000. | +/- | -NaN |
| 4.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.09 | +/- | 0. |
| 2.09 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 17.10 | +/- | 0. |
| 1.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.77 |
| -9999.00 |