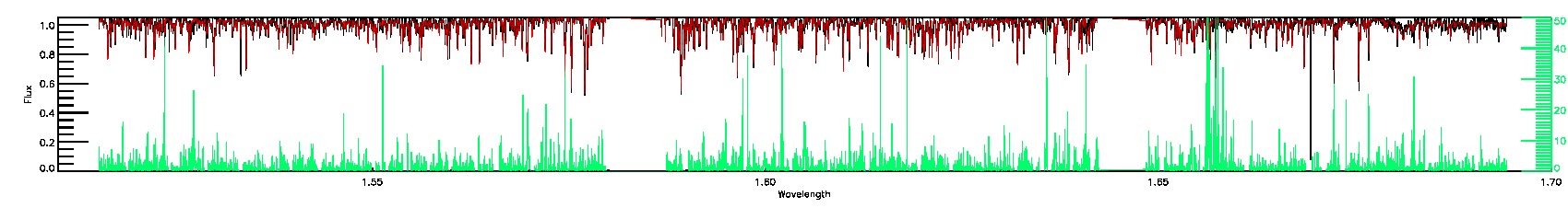
175-27-O
| 5609. | +/- | 12. |
| 5574. | +/- | 115. |
| 3.93 | +/- | 0. |
| 3.91 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |

175-27-O
| 5497. | +/- | 17. |
| 5465. | +/- | 127. |
| 4.40 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.57 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.11 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 9.46 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.00 |
| -9999.00 |
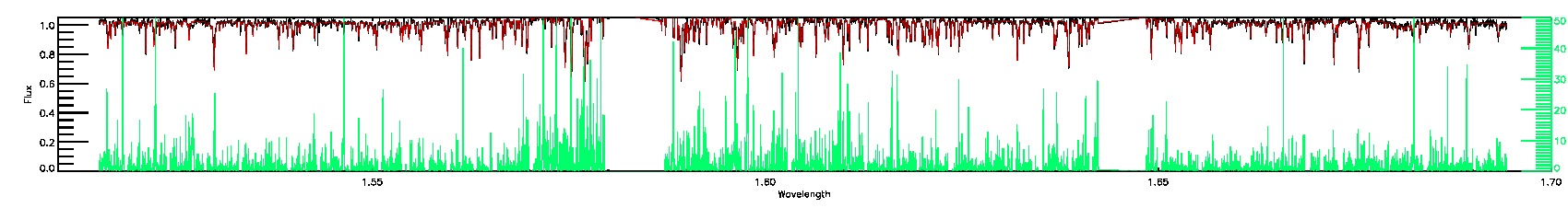
175-27-O
| 5029. | +/- | 17. |
| 5075. | +/- | 121. |
| 3.20 | +/- | 0. |
| 3.08 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 3.81 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.62 |
| -9999.00 |
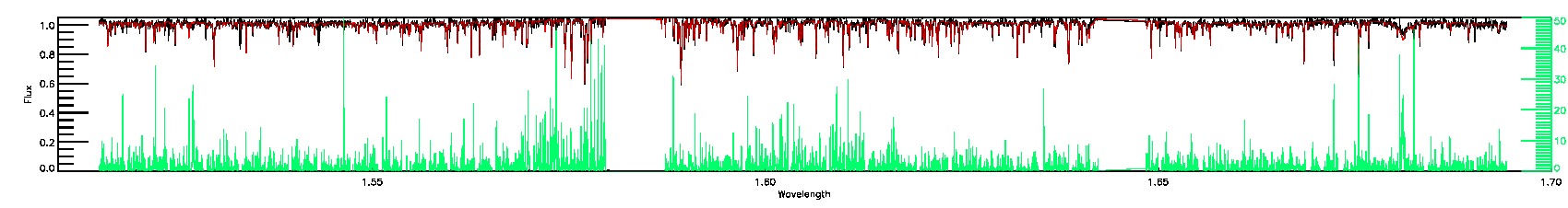
STAR_WARN,SN_WARN
175-27-O
| 4720. | +/- | 20. |
| 4793. | +/- | 120. |
| 2.73 | +/- | 0. |
| 2.54 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.79 |
| -9999.00 |
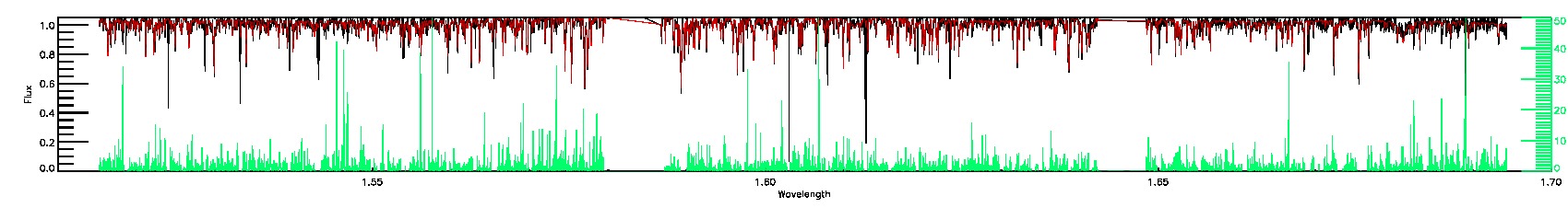
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
175-27-O
| 5637. | +/- | 81. |
| -10000. | +/- | -NaN |
| 4.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.58 | +/- | 0. |
| 0.58 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.79 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.16 |
| -9999.00 |

175-27-O
| 4919. | +/- | 15. |
| 4964. | +/- | 117. |
| 4.56 | +/- | 0. |
| 4.68 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.04 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.61 |
| -9999.00 |

175-27-O
| 5778. | +/- | 29. |
| 5720. | +/- | 153. |
| 4.40 | +/- | 0. |
| 4.33 | +/- | 0. |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.09 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.23 |
| -9999.00 |

175-27-O
| 5585. | +/- | 16. |
| 5548. | +/- | 131. |
| 4.50 | +/- | 0. |
| 4.45 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.84 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.49 |
| -9999.00 |

175-27-O
| 5499. | +/- | 20. |
| 5473. | +/- | 132. |
| 4.52 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 8.53 | +/- | 0. |
| 0.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 4384. | +/- | 6. |
| 4479. | +/- | 103. |
| 4.25 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.68 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.43 |
| -9999.00 |

SUSPECT_BROAD_LINES
175-27-O
| 6456. | +/- | 20. |
| 6331. | +/- | 146. |
| 4.24 | +/- | 0. |
| 4.15 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.14 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 66.54 | +/- | 0. |
| 1.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.11 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 4042. | +/- | 12. |
| 4126. | +/- | 97. |
| 4.46 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.53 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 1.52 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.23 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 4574. | +/- | 12. |
| 4646. | +/- | 109. |
| 4.44 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.09 | +/- | 0. |
| 2.78 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.33 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
175-27-O
| 3859. | +/- | 7. |
| 3958. | +/- | 89. |
| 4.43 | +/- | 0. |
| 4.80 | +/- | 0. |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.92 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.02 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN,SN_WARN
175-27-O
| 3375. | +/- | 9. |
| 3472. | +/- | 81. |
| 4.73 | +/- | 0. |
| 5.17 | +/- | 0. |
| 2.19 | +/- | 0. |
| 2.19 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 10.57 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.49 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 4632. | +/- | 12. |
| 4726. | +/- | 116. |
| 2.79 | +/- | 0. |
| 2.64 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.21 | +/- | 0. |
| 3.91 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.07 |
| -9999.00 |
175-27-O
| 6039. | +/- | 26. |
| 5952. | +/- | 150. |
| 4.18 | +/- | 0. |
| 4.07 | +/- | 0. |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 7.89 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.94 |
| -9999.00 |
STAR_WARN,COLORTE_WARN,SN_WARN
175-27-O
| 6439. | +/- | 67. |
| 6319. | +/- | 200. |
| 4.23 | +/- | 0. |
| 4.18 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.13 | +/- | 0. |
| 5.88 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.46 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 4611. | +/- | 13. |
| 4685. | +/- | 112. |
| 4.48 | +/- | 0. |
| 4.58 | +/- | 0. |
| 0.74 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.58 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.18 |
| -9999.00 |
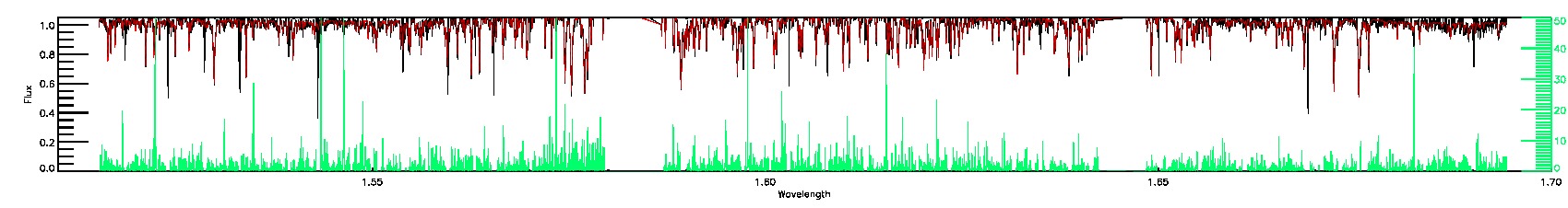
STAR_WARN,SN_WARN
175-27-O
| 5945. | +/- | 72. |
| 5886. | +/- | 172. |
| 4.39 | +/- | 0. |
| 4.46 | +/- | 0. |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.91 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.56 |
| -9999.00 |
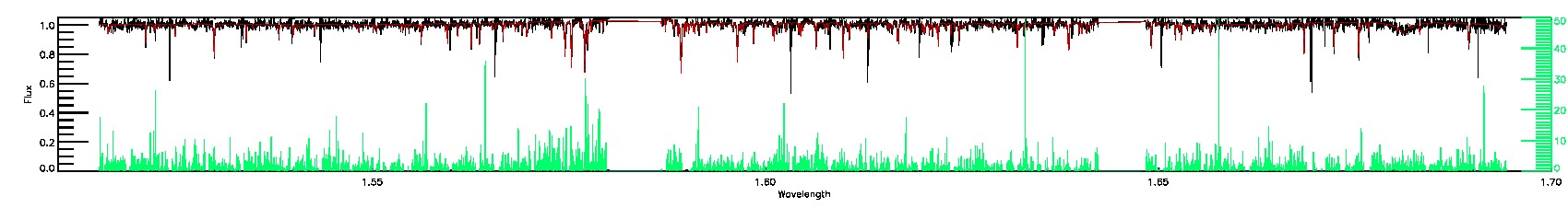
BRIGHT_NEIGHBOR
175-27-O
| 5364. | +/- | 23. |
| 5354. | +/- | 134. |
| 4.68 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.99 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.29 |
| -9999.00 |

175-27-O
| 5879. | +/- | 29. |
| 5820. | +/- | 152. |
| 4.31 | +/- | 0. |
| 4.32 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.16 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.08 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 3794. | +/- | 5. |
| 3887. | +/- | 84. |
| 4.46 | +/- | 0. |
| 4.77 | +/- | 0. |
| 0.54 | +/- | 0. |
| 0.54 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.98 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
175-27-O
| 5430. | +/- | 8. |
| 5399. | +/- | 110. |
| 4.38 | +/- | 0. |
| 4.24 | +/- | 0. |
| 0.59 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.26 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.04 | +/- | 0. |
| 1.66 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.28 |
| -9999.00 |

175-27-O
| 5411. | +/- | 41. |
| 5417. | +/- | 145. |
| 4.36 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -0.54 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.25 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -0.72 |
| -9999.00 |
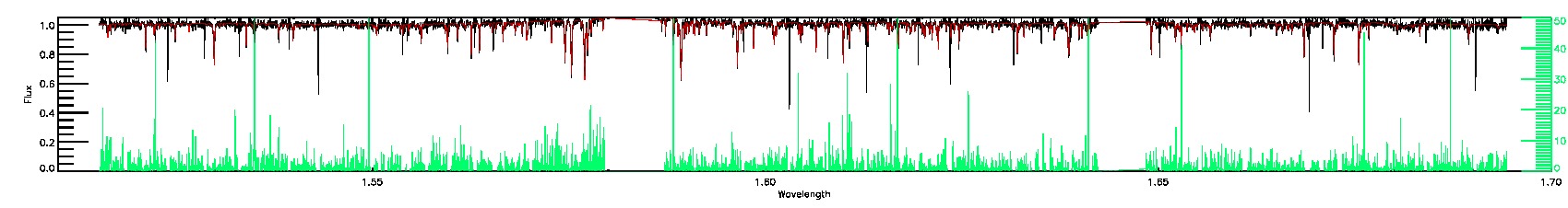
STAR_WARN,SN_WARN
175-27-O
| 6185. | +/- | 82. |
| 6091. | +/- | 189. |
| 4.12 | +/- | 0. |
| 4.07 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.03 | +/- | 1. |
| 0.03 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.10 | +/- | 0. |
| 6.35 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.35 |
| -9999.00 |
175-27-O
| 4813. | +/- | 13. |
| 4883. | +/- | 117. |
| 2.57 | +/- | 0. |
| 2.31 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.80 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.35 |
| -9999.00 |

175-27-O
| 5825. | +/- | 24. |
| 5762. | +/- | 150. |
| 4.34 | +/- | 0. |
| 4.27 | +/- | 0. |
| 1.22 | +/- | 0. |
| 1.22 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 1.67 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.05 |
| -9999.00 |

SUSPECT_BROAD_LINES
175-27-O
| 7462. | +/- | 11. |
| 7259. | +/- | 189. |
| 4.38 | +/- | 0. |
| 3.96 | +/- | 0. |
| 3.53 | +/- | 0. |
| 3.53 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.12 | +/- | 0. |
| 17.67 | +/- | 0. |
| 1.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.25 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 6374. | +/- | 58. |
| 6257. | +/- | 189. |
| 4.57 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 7.87 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.50 |
| -9999.00 |
175-27-O
| 4950. | +/- | 7. |
| 4981. | +/- | 99. |
| 4.43 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 3.69 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.04 |
| -9999.00 |

175-27-O
| 5148. | +/- | 19. |
| 5165. | +/- | 127. |
| 4.46 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 1.89 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.07 |
| -9999.00 |

175-27-O
| 5991. | +/- | 12. |
| 5898. | +/- | 120. |
| 4.36 | +/- | 0. |
| 4.15 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.01 |
| -9999.00 |
175-27-O
| 4526. | +/- | 5. |
| 4620. | +/- | 97. |
| 2.55 | +/- | 0. |
| 2.35 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.31 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.08 |
| -9999.00 |

STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN
175-27-O
| 4828. | +/- | 17. |
| -10000. | +/- | -NaN |
| 3.57 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| -0.83 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.62 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.89 |
| -9999.00 |
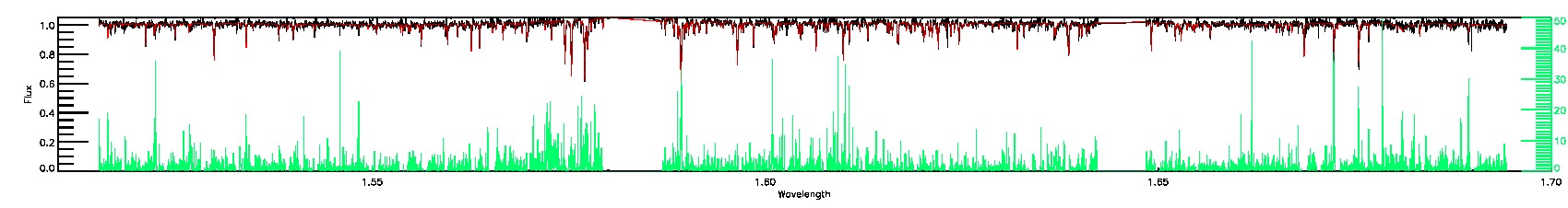
175-27-O
| 5552. | +/- | 18. |
| 5521. | +/- | 130. |
| 3.92 | +/- | 0. |
| 3.88 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.06 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.01 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5357. | +/- | 39. |
| 5378. | +/- | 148. |
| 3.84 | +/- | 0. |
| 4.03 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| -0.76 | +/- | 0. |
| -0.76 | +/- | 0. |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.41 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.01 |
| -9999.00 |
175-27-O
| 5608. | +/- | 13. |
| 5560. | +/- | 125. |
| 4.34 | +/- | 0. |
| 4.21 | +/- | 0. |
| 0.86 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.79 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.00 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 3256. | +/- | 5. |
| 3363. | +/- | 72. |
| 4.49 | +/- | 0. |
| 5.05 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 11.34 | +/- | 0. |
| 1.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
175-27-O
| 5944. | +/- | 26. |
| 5870. | +/- | 155. |
| 4.39 | +/- | 0. |
| 4.31 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.08 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5376. | +/- | 34. |
| 5353. | +/- | 142. |
| 4.46 | +/- | 0. |
| 4.35 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 1.55 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.22 |
| -9999.00 |
175-27-O
| 4574. | +/- | 9. |
| 4664. | +/- | 109. |
| 2.89 | +/- | 0. |
| 2.71 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.17 | +/- | 0. |
| 3.37 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.32 |
| -9999.00 |
175-27-O
| 4640. | +/- | 7. |
| 4707. | +/- | 98. |
| 2.96 | +/- | 0. |
| 2.75 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.74 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.22 |
| -9999.00 |

175-27-O
| 5388. | +/- | 18. |
| 5367. | +/- | 131. |
| 4.50 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.48 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.15 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 1.66 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.28 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5302. | +/- | 44. |
| 5309. | +/- | 143. |
| 4.26 | +/- | 0. |
| 4.37 | +/- | 0. |
| 0.61 | +/- | 0. |
| 0.61 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.49 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 1.00 |
| -9999.00 |
175-27-O
| 4777. | +/- | 8. |
| 4829. | +/- | 104. |
| 4.55 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.51 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.76 |
| -9999.00 |

175-27-O
| 4794. | +/- | 5. |
| 4857. | +/- | 88. |
| 2.72 | +/- | 0. |
| 2.40 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.33 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.27 |
| -9999.00 |

175-27-O
| 4817. | +/- | 14. |
| 4878. | +/- | 114. |
| 3.08 | +/- | 0. |
| 2.92 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.05 |
| -9999.00 |

175-27-O
| 5053. | +/- | 12. |
| 5086. | +/- | 114. |
| 2.79 | +/- | 0. |
| 2.45 | +/- | 0. |
| 1.89 | +/- | 0. |
| 1.89 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.33 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.29 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
175-27-O
| 4244. | +/- | 3. |
| 4329. | +/- | 85. |
| 4.41 | +/- | 0. |
| 4.57 | +/- | 0. |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 4.25 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.44 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5985. | +/- | 72. |
| 5912. | +/- | 176. |
| 4.25 | +/- | 0. |
| 4.23 | +/- | 0. |
| 0.48 | +/- | 0. |
| 0.48 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.30 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5918. | +/- | 50. |
| 5857. | +/- | 169. |
| 4.14 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.58 | +/- | 0. |
| 0.58 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.35 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 1.84 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
175-27-O
| 5090. | +/- | 8. |
| 5131. | +/- | 99. |
| 3.79 | +/- | 0. |
| 3.89 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -0.49 | +/- | 0. |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.21 | +/- | 0. |
| 3.94 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.24 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
175-27-O
| 3901. | +/- | 4. |
| 4000. | +/- | 83. |
| 4.43 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.66 | +/- | 0. |
| 0.66 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.85 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
175-27-O
| 4938. | +/- | 5. |
| 4967. | +/- | 87. |
| 4.56 | +/- | 0. |
| 4.53 | +/- | 0. |
| 0.91 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 1.52 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.22 |
| -9999.00 |

175-27-O
| 4432. | +/- | 5. |
| 4527. | +/- | 96. |
| 4.19 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 11.54 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.24 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5627. | +/- | 31. |
| 5580. | +/- | 147. |
| 4.34 | +/- | 0. |
| 4.23 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.10 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.38 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 4303. | +/- | 6. |
| 4407. | +/- | 99. |
| 4.26 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.23 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.55 |
| -9999.00 |

175-27-O
| 6128. | +/- | 45. |
| 6041. | +/- | 169. |
| 4.30 | +/- | 0. |
| 4.27 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.15 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 6.60 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.99 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 6058. | +/- | 51. |
| 5976. | +/- | 170. |
| 4.33 | +/- | 0. |
| 4.27 | +/- | 0. |
| 0.47 | +/- | 0. |
| 0.47 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 14.08 | +/- | 0. |
| 1.15 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.18 |
| -9999.00 |
175-27-O
| 5067. | +/- | 12. |
| 5093. | +/- | 114. |
| 4.56 | +/- | 0. |
| 4.63 | +/- | 0. |
| 1.92 | +/- | 0. |
| 1.92 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.86 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.82 |
| -9999.00 |
SUSPECT_BROAD_LINES
175-27-O
| 6510. | +/- | 16. |
| 6362. | +/- | 142. |
| 4.18 | +/- | 0. |
| 3.92 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.68 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 40.59 | +/- | 0. |
| 1.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
175-27-O
| 5554. | +/- | 24. |
| 5530. | +/- | 139. |
| 3.94 | +/- | 0. |
| 3.97 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | 0. |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.37 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.40 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD
175-27-O
| 4504. | +/- | 11. |
| -10000. | +/- | -NaN |
| 3.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 95.96 | +/- | 0. |
| 1.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.06 |
| -9999.00 |

STAR_BAD
175-27-O
| 5967. | +/- | 47. |
| -10000. | +/- | -NaN |
| 4.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.11 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.12 |
| -9999.00 |

175-27-O
| 5172. | +/- | 8. |
| 5184. | +/- | 97. |
| 4.62 | +/- | 0. |
| 4.66 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.87 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |

175-27-O
| 5010. | +/- | 20. |
| 5051. | +/- | 125. |
| 3.55 | +/- | 0. |
| 3.46 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.48 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.36 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
175-27-O
| 3603. | +/- | 7. |
| 3694. | +/- | 84. |
| 4.52 | +/- | 0. |
| 4.86 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -0.41 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.49 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 4184. | +/- | 8. |
| 4268. | +/- | 97. |
| 4.36 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 5.05 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.52 |
| -9999.00 |

175-27-O
| 5725. | +/- | 22. |
| 5666. | +/- | 138. |
| 4.22 | +/- | 0. |
| 4.09 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.19 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.11 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.18 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 1.00 |
| -9999.00 |

175-27-O
| 4683. | +/- | 7. |
| 4754. | +/- | 99. |
| 4.46 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.58 | +/- | 0. |
| 0.58 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.52 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.84 |
| -9999.00 |

175-27-O
| 4482. | +/- | 8. |
| 4586. | +/- | 106. |
| 3.07 | +/- | 0. |
| 2.93 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.68 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.63 |
| -9999.00 |
175-27-O
| 4266. | +/- | 7. |
| 4356. | +/- | 97. |
| 4.13 | +/- | 0. |
| 4.33 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 12.22 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.16 |
| -9999.00 |

175-27-O
| 4782. | +/- | 10. |
| 4849. | +/- | 106. |
| 2.87 | +/- | 0. |
| 2.70 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.44 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.07 |
| -9999.00 |

175-27-O
| 4646. | +/- | 8. |
| 4704. | +/- | 103. |
| 3.19 | +/- | 0. |
| 2.97 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.47 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.41 |
| -9999.00 |

175-27-O
| 4749. | +/- | 15. |
| 4828. | +/- | 118. |
| 2.54 | +/- | 0. |
| 2.30 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.86 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |

175-27-O
| 5124. | +/- | 17. |
| 5154. | +/- | 123. |
| 3.54 | +/- | 0. |
| 3.45 | +/- | 0. |
| 1.21 | +/- | 0. |
| 1.21 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.56 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.02 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
175-27-O
| 5370. | +/- | 98. |
| -10000. | +/- | -NaN |
| 4.58 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.85 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 1.00 |
| -9999.00 |

SUSPECT_BROAD_LINES
175-27-O
| 6763. | +/- | 22. |
| 6601. | +/- | 175. |
| 4.19 | +/- | 0. |
| 4.03 | +/- | 0. |
| 0.30 | +/- | 0. |
| 0.30 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.21 | +/- | 0. |
| 49.16 | +/- | 0. |
| 1.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.79 |
| -9999.00 |
175-27-O
| 5344. | +/- | 12. |
| 5327. | +/- | 118. |
| 4.07 | +/- | 0. |
| 3.99 | +/- | 0. |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.31 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.57 |
| -9999.00 |

175-27-O
| 4021. | +/- | 5. |
| 4114. | +/- | 86. |
| 4.47 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.56 | +/- | 0. |
| 0.56 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.25 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.99 |
| -9999.00 |
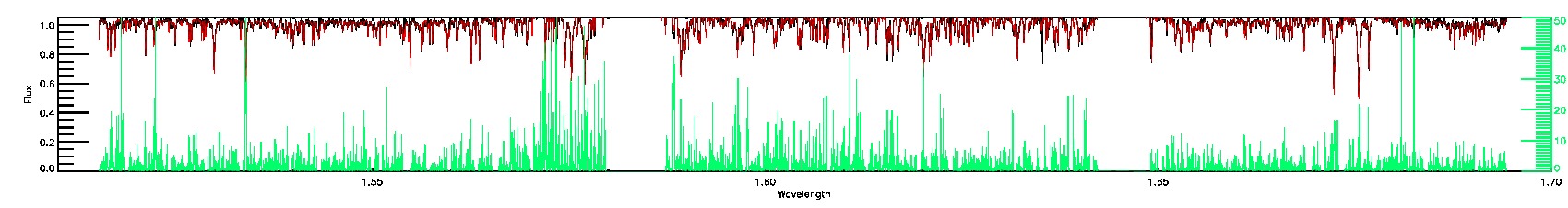
175-27-O
| 4598. | +/- | 7. |
| 4685. | +/- | 107. |
| 4.40 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.95 | +/- | 0. |
| 0.95 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.13 | +/- | 0. |
| 1.55 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |

175-27-O
| 6050. | +/- | 49. |
| 5978. | +/- | 172. |
| 3.98 | +/- | 0. |
| 4.02 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 5.58 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.98 |
| -9999.00 |
175-27-O
| 6044. | +/- | 15. |
| 5955. | +/- | 125. |
| 4.54 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.64 | +/- | 0. |
| 0.64 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.73 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
BRIGHT_NEIGHBOR
175-27-O
| 4169. | +/- | 5. |
| 4260. | +/- | 93. |
| 4.46 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.92 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.42 |
| -9999.00 |

175-27-O
| 5838. | +/- | 13. |
| 5774. | +/- | 122. |
| 4.49 | +/- | 0. |
| 4.41 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.34 |
| -9999.00 |

175-27-O
| 4188. | +/- | 1. |
| 4296. | +/- | 76. |
| 1.76 | +/- | 0. |
| 1.60 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.85 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.43 |
| -9999.00 |

175-27-O
| 5442. | +/- | 20. |
| 5420. | +/- | 133. |
| 4.45 | +/- | 0. |
| 4.41 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 1.94 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.21 |
| -9999.00 |

175-27-O
| 5054. | +/- | 12. |
| 5071. | +/- | 115. |
| 4.45 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 2.33 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |

175-27-O
| 5891. | +/- | 13. |
| 5809. | +/- | 134. |
| 4.30 | +/- | 0. |
| 4.10 | +/- | 0. |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.93 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.20 |
| -9999.00 |
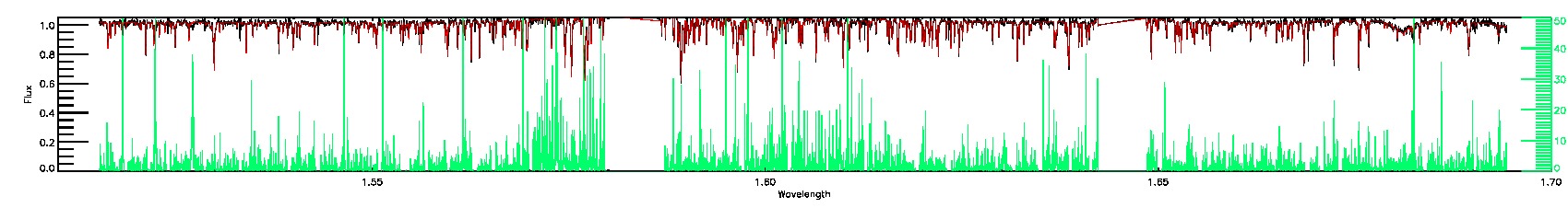
175-27-O
| 4836. | +/- | 9. |
| 4898. | +/- | 104. |
| 3.66 | +/- | 0. |
| 3.65 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.17 | +/- | 0. |
| 3.52 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.01 |
| -9999.00 |

175-27-O
| 4817. | +/- | 7. |
| 4889. | +/- | 91. |
| 2.97 | +/- | 0. |
| 2.84 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -0.47 | +/- | 0. |
| -0.47 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.90 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.38 |
| -9999.00 |

175-27-O
| 4740. | +/- | 5. |
| 4792. | +/- | 89. |
| 3.36 | +/- | 0. |
| 3.16 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.62 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.96 |
| -9999.00 |
| 0.16 |
| -9999.00 |

175-27-O
| 5331. | +/- | 12. |
| 5317. | +/- | 117. |
| 4.67 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.93 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.02 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.97 |
| -9999.00 |
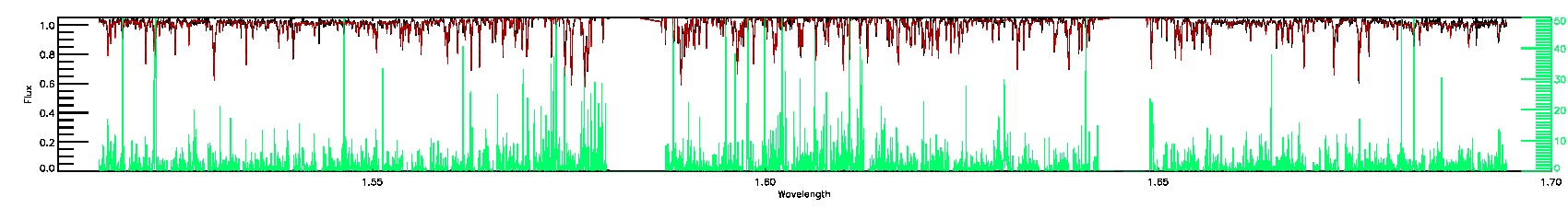
175-27-O
| 4603. | +/- | 3. |
| 4665. | +/- | 78. |
| 2.83 | +/- | 0. |
| 2.57 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.19 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| 2.42 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.25 |
| -9999.00 |
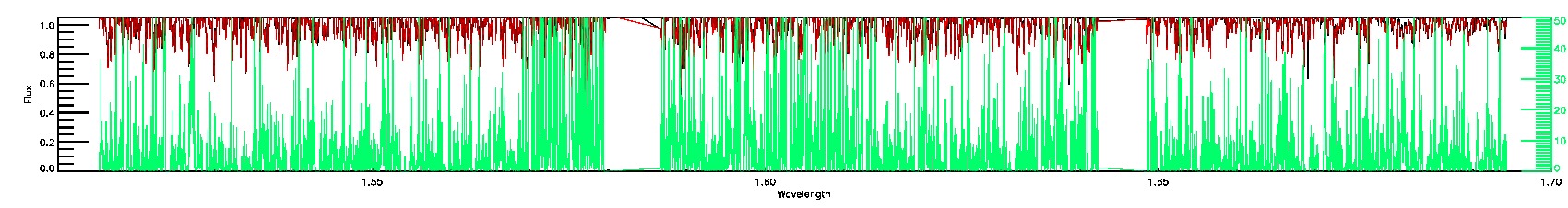
STAR_WARN,SN_WARN
175-27-O
| 4701. | +/- | 12. |
| 4785. | +/- | 117. |
| 4.37 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.17 | +/- | 0. |
| 2.40 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.32 |
| -9999.00 |

175-27-O
| 4582. | +/- | 4. |
| 4667. | +/- | 84. |
| 2.66 | +/- | 0. |
| 2.46 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 3.23 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.03 |
| -9999.00 |

STAR_BAD
STAR_WARN,COLORTE_WARN
175-27-O
| 4211. | +/- | 7. |
| -10000. | +/- | -NaN |
| 4.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.68 | +/- | 0. |
| 0.68 | +/- | -NaN |
| -0.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.02 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -2.01 |
| -9999.00 |
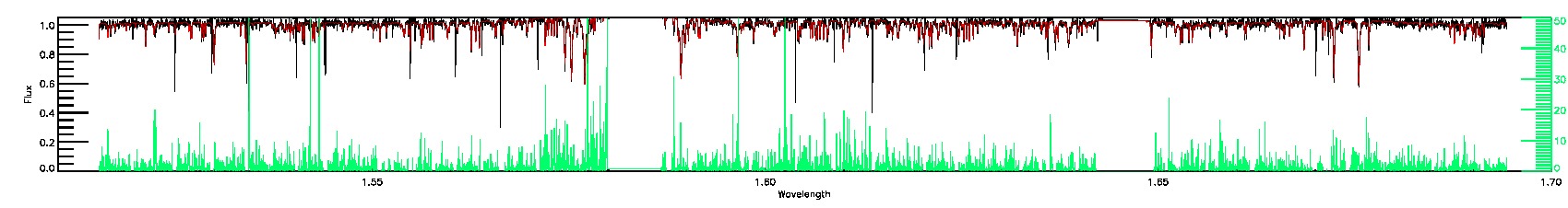
175-27-O
| 4933. | +/- | 7. |
| 4989. | +/- | 99. |
| 2.75 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.65 | +/- | 0. |
| 1.65 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.77 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.23 |
| -9999.00 |

175-27-O
| 5401. | +/- | 21. |
| 5385. | +/- | 134. |
| 4.49 | +/- | 0. |
| 4.47 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.70 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.47 |
| -9999.00 |

175-27-O
| 5968. | +/- | 39. |
| 5912. | +/- | 165. |
| 3.97 | +/- | 0. |
| 4.07 | +/- | 0. |
| 0.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| -0.60 | +/- | 0. |
| -0.60 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 5.12 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.24 |
| -9999.00 |
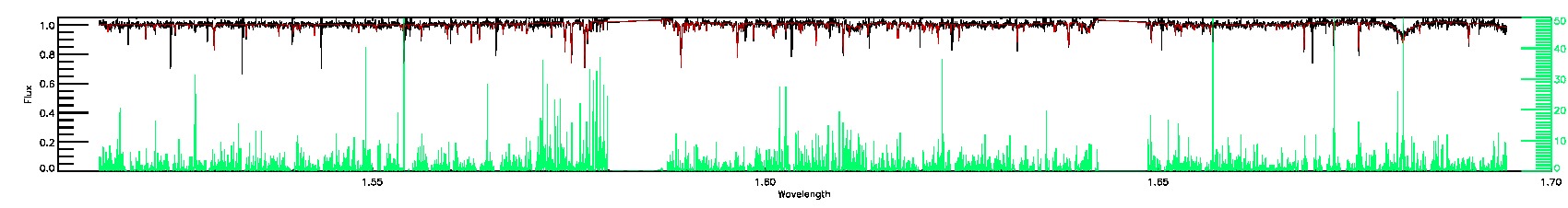
175-27-O
| 5038. | +/- | 19. |
| 5078. | +/- | 123. |
| 3.83 | +/- | 0. |
| 3.91 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.15 | +/- | 0. |
| 1.55 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 6089. | +/- | 88. |
| 6016. | +/- | 186. |
| 4.36 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| -0.54 | +/- | 0. |
| -0.53 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.20 | +/- | 0. |
| 9.25 | +/- | 0. |
| 0.97 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.49 |
| -9999.00 |
175-27-O
| 4775. | +/- | 6. |
| 4840. | +/- | 91. |
| 3.14 | +/- | 0. |
| 2.99 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.34 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.37 |
| -9999.00 |

175-27-O
| 4441. | +/- | 6. |
| 4537. | +/- | 102. |
| 4.34 | +/- | 0. |
| 4.57 | +/- | 0. |
| 0.62 | +/- | 0. |
| 0.62 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 10.29 | +/- | 0. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.18 |
| -9999.00 |

175-27-O
| 4424. | +/- | 2. |
| 4506. | +/- | 76. |
| 4.51 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.52 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 1.90 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.17 |
| -9999.00 |

175-27-O
| 4734. | +/- | 5. |
| 4791. | +/- | 91. |
| 3.42 | +/- | 0. |
| 3.24 | +/- | 0. |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.78 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.12 |
| -9999.00 |

175-27-O
| 4780. | +/- | 12. |
| 4836. | +/- | 111. |
| 4.62 | +/- | 0. |
| 4.71 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| -0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.28 |
| -9999.00 |

175-27-O
| 6391. | +/- | 14. |
| 6255. | +/- | 136. |
| 4.53 | +/- | 0. |
| 4.27 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.43 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5939. | +/- | 48. |
| 5859. | +/- | 167. |
| 4.44 | +/- | 0. |
| 4.29 | +/- | 0. |
| 0.59 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.15 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.55 |
| -9999.00 |

175-27-O
| 4561. | +/- | 3. |
| 4643. | +/- | 80. |
| 2.67 | +/- | 0. |
| 2.37 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.02 | +/- | 0. |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.13 | +/- | 0. |
| 3.00 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -0.18 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 3578. | +/- | 5. |
| 3661. | +/- | 78. |
| 4.58 | +/- | 0. |
| 4.86 | +/- | 0. |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 3.57 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.24 |
| -9999.00 |
175-27-O
| 5578. | +/- | 18. |
| 5546. | +/- | 128. |
| 4.51 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.66 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.31 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5765. | +/- | 33. |
| 5716. | +/- | 156. |
| 4.43 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.63 | +/- | 0. |
| 0.63 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.70 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.10 |
| -9999.00 |
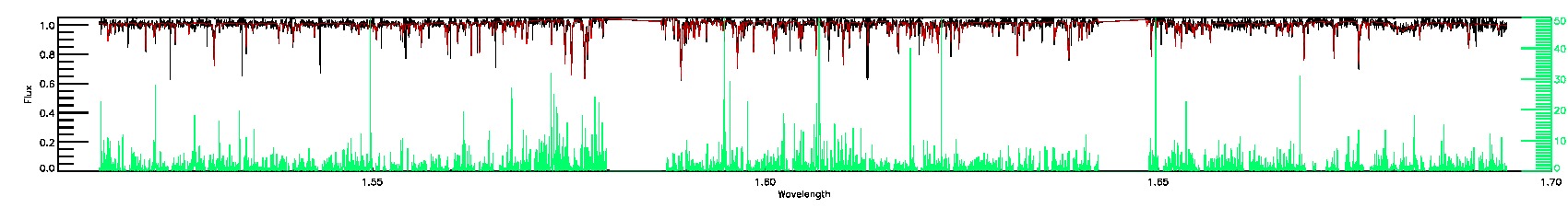
175-27-O
| 5934. | +/- | 29. |
| 5865. | +/- | 155. |
| 4.54 | +/- | 0. |
| 4.50 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.26 | +/- | 0. |
| 0.35 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.10 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 4510. | +/- | 9. |
| 4600. | +/- | 107. |
| 4.45 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.70 | +/- | 0. |
| 0.70 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.80 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.32 |
| -9999.00 |

175-27-O
| 5833. | +/- | 19. |
| 5768. | +/- | 140. |
| 4.57 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.34 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.76 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 4754. | +/- | 19. |
| 4810. | +/- | 117. |
| 3.35 | +/- | 0. |
| 3.18 | +/- | 0. |
| 1.12 | +/- | 0. |
| 1.12 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.87 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.25 |
| -9999.00 |
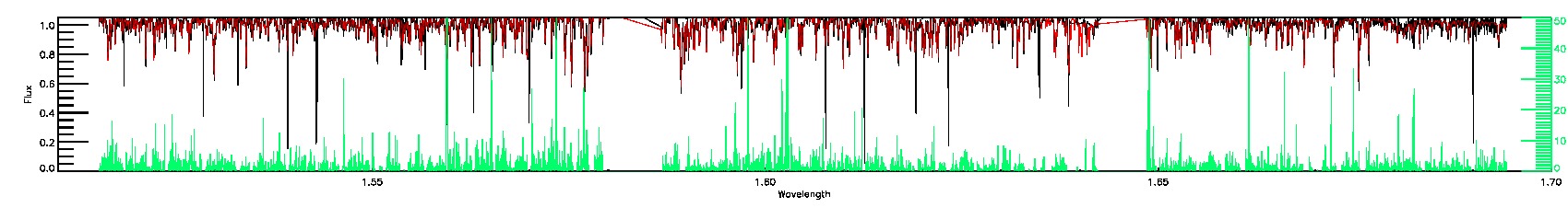
STAR_WARN,SN_WARN
175-27-O
| 4860. | +/- | 23. |
| 4923. | +/- | 124. |
| 4.47 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -0.40 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 2.87 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.64 |
| -9999.00 |
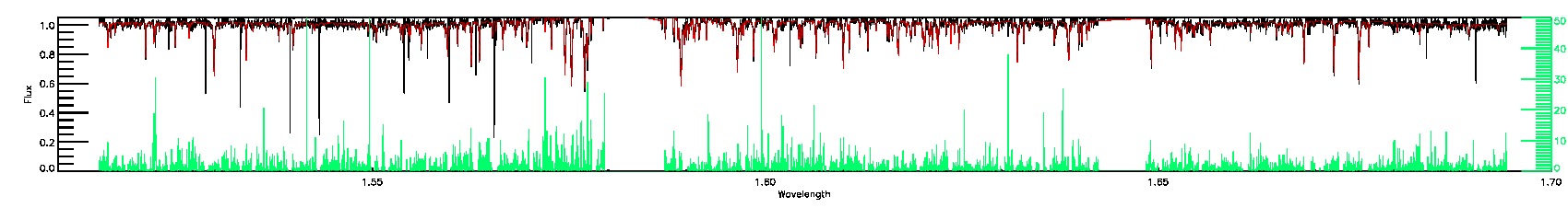
175-27-O
| 4657. | +/- | 4. |
| 4726. | +/- | 83. |
| 2.89 | +/- | 0. |
| 2.68 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.94 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.47 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 3677. | +/- | 2. |
| 3781. | +/- | 66. |
| 4.44 | +/- | 0. |
| 4.89 | +/- | 0. |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| -0.36 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.52 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.28 |
| -9999.00 |
SUSPECT_BROAD_LINES
175-27-O
| 6601. | +/- | 16. |
| 6446. | +/- | 147. |
| 4.37 | +/- | 0. |
| 4.12 | +/- | 0. |
| 1.86 | +/- | 0. |
| 1.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 23.12 | +/- | 0. |
| 1.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.27 |
| -9999.00 |
SUSPECT_BROAD_LINES
STAR_WARN,SN_WARN
175-27-O
| 5886. | +/- | 54. |
| 5835. | +/- | 171. |
| 4.32 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.85 | +/- | 0. |
| 0.85 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 21.10 | +/- | 0. |
| 1.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.75 |
| -9999.00 |
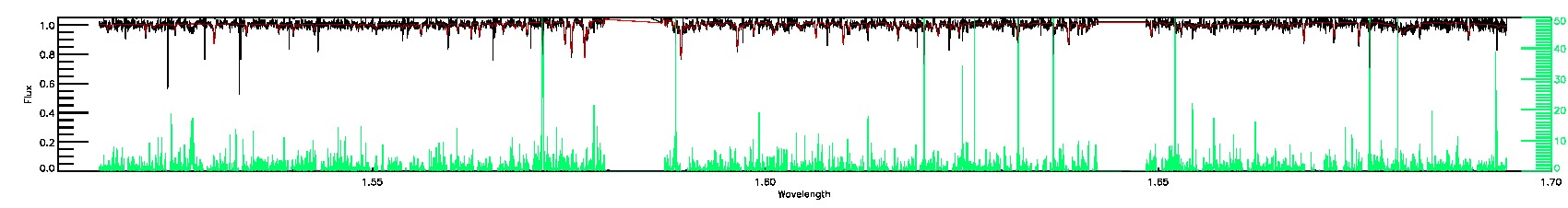
SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN
175-27-O
| 3422. | +/- | 4. |
| 3509. | +/- | 71. |
| 4.58 | +/- | 0. |
| 4.91 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 20.26 | +/- | 0. |
| 1.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.27 |
| -9999.00 |
175-27-O
| 6188. | +/- | 34. |
| 6083. | +/- | 161. |
| 4.46 | +/- | 0. |
| 4.32 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 10.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
175-27-O
| 5782. | +/- | 19. |
| 5725. | +/- | 139. |
| 4.43 | +/- | 0. |
| 4.37 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.50 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
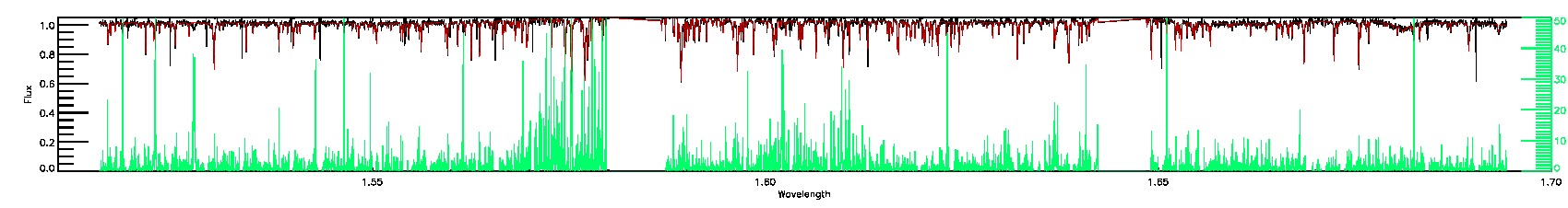
175-27-O
| 5575. | +/- | 27. |
| 5546. | +/- | 143. |
| 4.48 | +/- | 0. |
| 4.50 | +/- | 0. |
| 1.09 | +/- | 0. |
| 1.09 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.80 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.99 |
| -9999.00 |
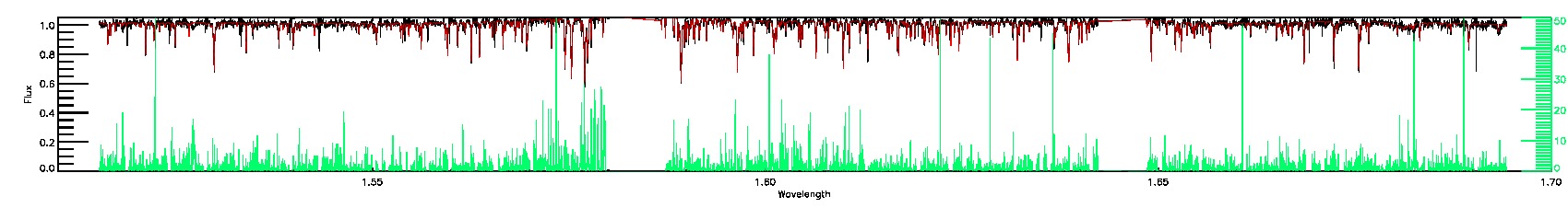
175-27-O
| 4659. | +/- | 6. |
| 4728. | +/- | 94. |
| 2.79 | +/- | 0. |
| 2.45 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.94 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.18 |
| -9999.00 |

175-27-O
| 5586. | +/- | 17. |
| 5568. | +/- | 125. |
| 4.41 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.98 | +/- | 0. |
| 0.98 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.15 | +/- | 0. |
| 1.80 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.36 |
| -9999.00 |
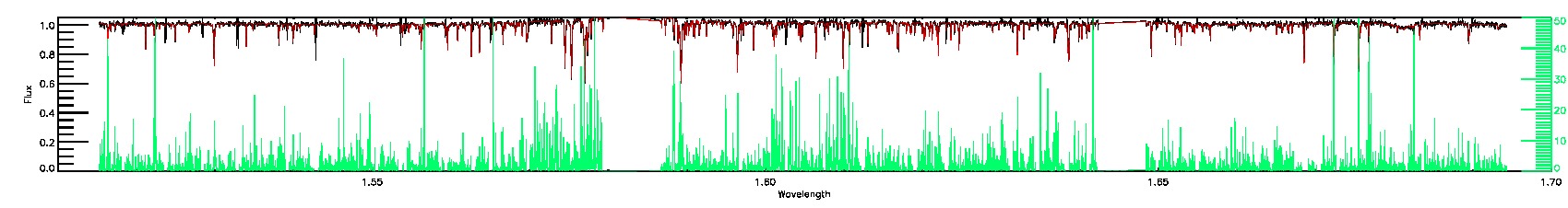
STAR_WARN,SN_WARN
175-27-O
| 4412. | +/- | 9. |
| 4515. | +/- | 108. |
| 2.21 | +/- | 0. |
| 2.02 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.53 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.08 | +/- | 0. |
| 3.62 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.25 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_WARN,COLORTE_WARN,SN_WARN
175-27-O
| 3620. | +/- | 8. |
| 3720. | +/- | 85. |
| 4.23 | +/- | 0. |
| 4.66 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 26.93 | +/- | 0. |
| 1.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.56 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 4841. | +/- | 26. |
| 4903. | +/- | 125. |
| 4.50 | +/- | 0. |
| 4.71 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.21 |
| -9999.00 |

SUSPECT_BROAD_LINES
175-27-O
| 4972. | +/- | 12. |
| 5000. | +/- | 109. |
| 4.54 | +/- | 0. |
| 4.54 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 17.48 | +/- | 0. |
| 1.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.14 |
| -9999.00 |
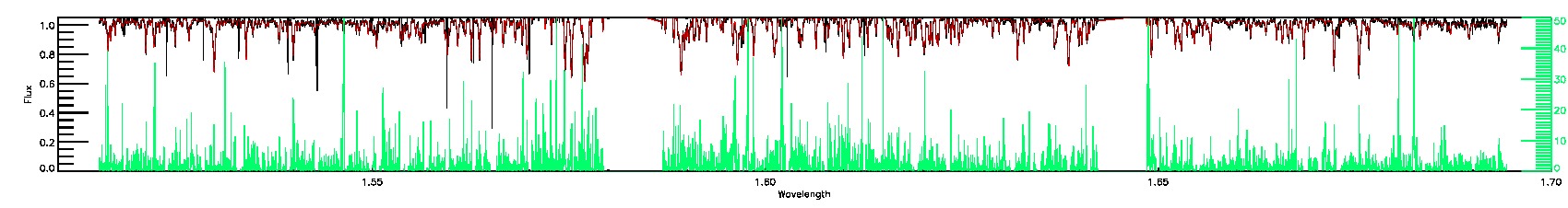
175-27-O
| 5817. | +/- | 13. |
| 5753. | +/- | 129. |
| 4.19 | +/- | 0. |
| 4.09 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.38 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.33 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.22 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
175-27-O
| 4063. | +/- | 9. |
| 4168. | +/- | 100. |
| 4.36 | +/- | 0. |
| 4.74 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.38 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.40 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.97 |
| -9999.00 |

175-27-O
| 4075. | +/- | 2. |
| 4188. | +/- | 89. |
| 1.58 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.82 | +/- | 0. |
| 1.82 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -0.56 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.11 | +/- | 0. |
| 0.61 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.41 |
| -9999.00 |

175-27-O
| 6159. | +/- | 42. |
| 6058. | +/- | 172. |
| 4.06 | +/- | 0. |
| 3.93 | +/- | 0. |
| 2.03 | +/- | 0. |
| 2.03 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 6.32 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.61 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5032. | +/- | 26. |
| 5088. | +/- | 134. |
| 3.59 | +/- | 0. |
| 3.59 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -0.67 | +/- | 0. |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -0.41 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.20 | +/- | 0. |
| 4.39 | +/- | 0. |
| 0.64 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.45 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.76 |
| -9999.00 |

175-27-O
| 5643. | +/- | 41. |
| 5610. | +/- | 151. |
| 4.40 | +/- | 0. |
| 4.44 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 6.00 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.61 |
| -9999.00 |
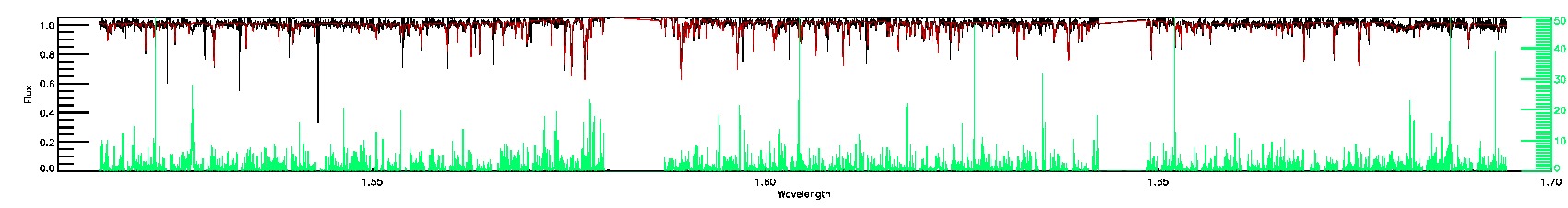
175-27-O
| 6246. | +/- | 35. |
| 6132. | +/- | 169. |
| 4.58 | +/- | 0. |
| 4.40 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 6.25 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.07 |
| -9999.00 |
175-27-O
| 6158. | +/- | 13. |
| 6049. | +/- | 127. |
| 4.57 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 5.75 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
175-27-O
| 4805. | +/- | 13. |
| 4867. | +/- | 115. |
| 4.52 | +/- | 0. |
| 4.69 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.36 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.19 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 1.68 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |

175-27-O
| 5468. | +/- | 14. |
| 5435. | +/- | 124. |
| 4.28 | +/- | 0. |
| 4.16 | +/- | 0. |
| 0.68 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.53 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 1.00 |
| -9999.00 |
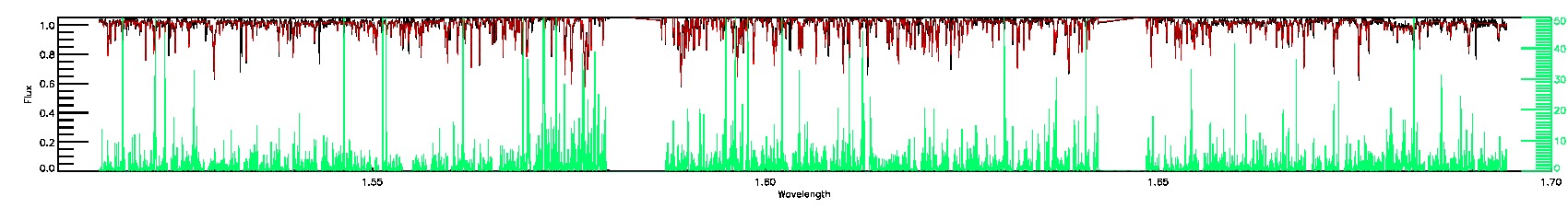
STAR_WARN,SN_WARN
175-27-O
| 4027. | +/- | 9. |
| 4133. | +/- | 99. |
| 4.40 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.41 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.24 |
| -9999.00 |

175-27-O
| 4362. | +/- | 2. |
| 4461. | +/- | 78. |
| 2.27 | +/- | 0. |
| 2.06 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.32 |
| -9999.00 |
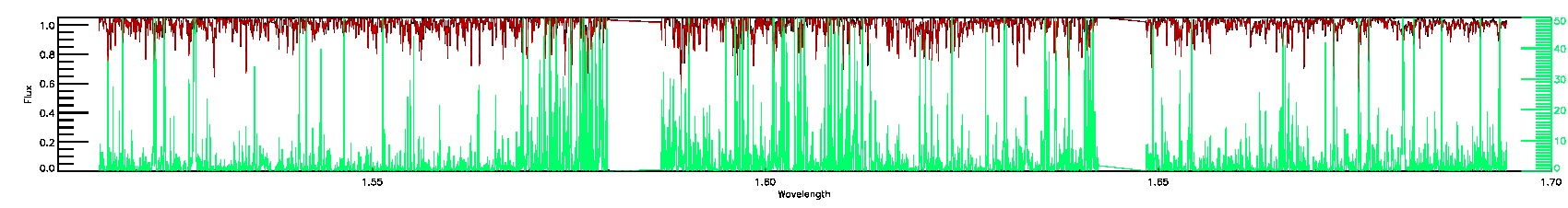
STAR_WARN,SN_WARN
175-27-O
| 5764. | +/- | 40. |
| 5707. | +/- | 154. |
| 4.30 | +/- | 0. |
| 4.22 | +/- | 0. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.35 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.08 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 4181. | +/- | 5. |
| 4274. | +/- | 95. |
| 4.36 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.85 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.16 |
| -9999.00 |

175-27-O
| 4918. | +/- | 14. |
| 4974. | +/- | 117. |
| 2.77 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.70 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.47 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 3994. | +/- | 4. |
| 4111. | +/- | 101. |
| 1.32 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.87 | +/- | 0. |
| 1.87 | +/- | -NaN |
| -0.69 | +/- | 0. |
| -0.69 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.04 |
| -9999.00 |
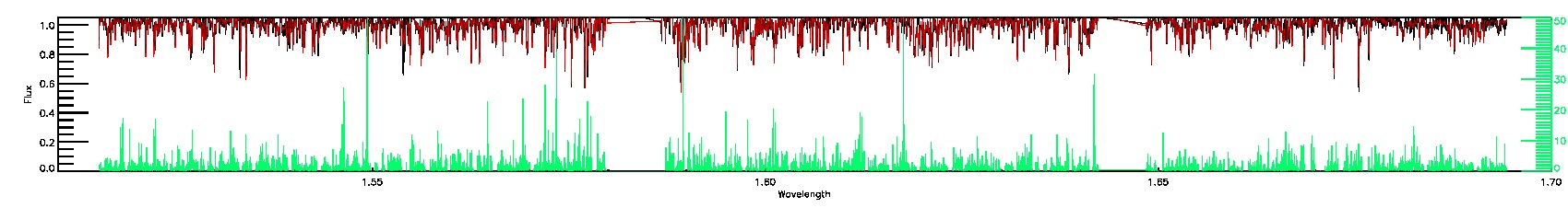
175-27-O
| 5328. | +/- | 13. |
| 5335. | +/- | 115. |
| 4.49 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.82 | +/- | 0. |
| 0.82 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -0.34 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.14 | +/- | 0. |
| 1.72 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.59 |
| -9999.00 |

175-27-O
| 5589. | +/- | 29. |
| 5571. | +/- | 145. |
| 3.86 | +/- | 0. |
| 3.94 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 3.91 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.46 |
| -9999.00 |

175-27-O
| 6032. | +/- | 36. |
| 5957. | +/- | 161. |
| 4.39 | +/- | 0. |
| 4.39 | +/- | 0. |
| 0.56 | +/- | 0. |
| 0.56 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.17 |
| -9999.00 |
SUSPECT_BROAD_LINES
175-27-O
| 6989. | +/- | 14. |
| 6800. | +/- | 170. |
| 4.52 | +/- | 0. |
| 4.31 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 14.79 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.08 |
| -9999.00 |
175-27-O
| 5858. | +/- | 15. |
| 5787. | +/- | 136. |
| 4.37 | +/- | 0. |
| 4.25 | +/- | 0. |
| 0.93 | +/- | 0. |
| 0.93 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.76 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |

BRIGHT_NEIGHBOR
175-27-O
| 5893. | +/- | 36. |
| 5841. | +/- | 156. |
| 4.42 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 5.10 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.20 |
| -9999.00 |

175-27-O
| 4774. | +/- | 12. |
| 4831. | +/- | 112. |
| 3.44 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 2.98 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.98 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5883. | +/- | 37. |
| 5814. | +/- | 161. |
| 4.46 | +/- | 0. |
| 4.38 | +/- | 0. |
| 1.26 | +/- | 0. |
| 1.26 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 6.27 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.46 |
| -9999.00 |

175-27-O
| 4200. | +/- | 1. |
| 4305. | +/- | 75. |
| 2.06 | +/- | 0. |
| 1.88 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.39 | +/- | 0. |
| -0.39 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.20 | +/- | 0. |
| 3.72 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.55 |
| -9999.00 |

175-27-O
| 5908. | +/- | 35. |
| 5839. | +/- | 159. |
| 4.37 | +/- | 0. |
| 4.31 | +/- | 0. |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.95 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.30 |
| -9999.00 |
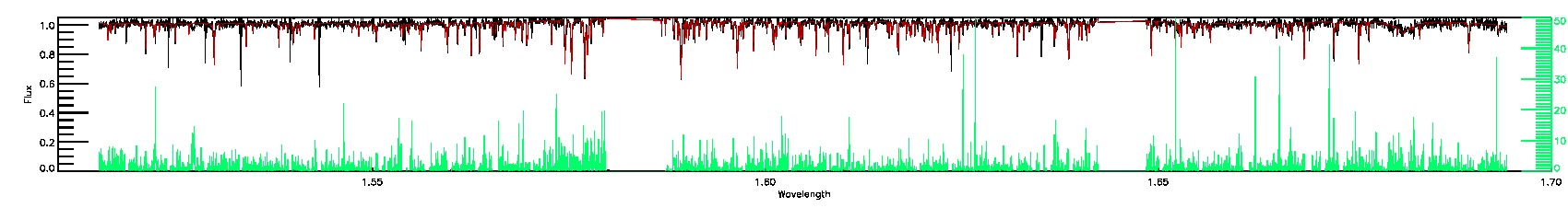
STAR_WARN,SN_WARN
175-27-O
| 4674. | +/- | 14. |
| 4764. | +/- | 117. |
| 2.60 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -0.51 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.98 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.31 |
| -9999.00 |
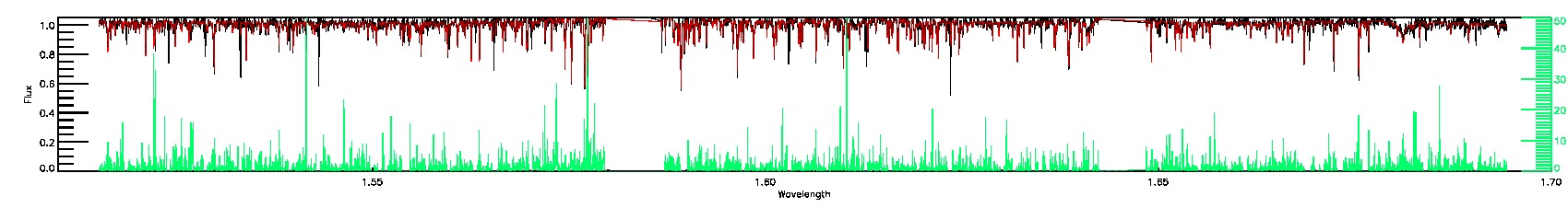
STAR_WARN,COLORTE_WARN
175-27-O
| 3976. | +/- | 3. |
| 4074. | +/- | 83. |
| 4.35 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.25 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.19 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
175-27-O
| 4833. | +/- | 127. |
| -10000. | +/- | -NaN |
| 4.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.48 | +/- | 0. |
| 1.48 | +/- | -NaN |
| -0.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 13.69 | +/- | 0. |
| 1.14 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.49 |
| -9999.00 |

175-27-O
| 5660. | +/- | 24. |
| 5627. | +/- | 142. |
| 4.50 | +/- | 0. |
| 4.56 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.20 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.15 | +/- | 0. |
| 1.70 | +/- | 0. |
| 0.23 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
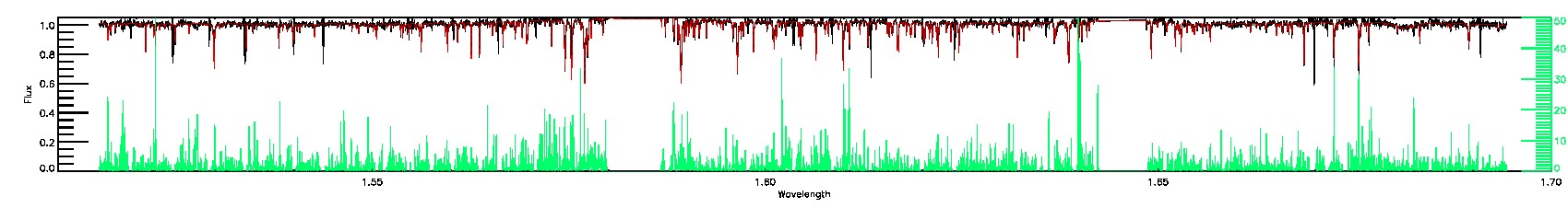
STAR_WARN,COLORTE_WARN
175-27-O
| 3893. | +/- | 4. |
| 3976. | +/- | 83. |
| 4.49 | +/- | 0. |
| 4.70 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.13 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.06 | +/- | 0. |
| 2.64 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 1.00 |
| -9999.00 |
175-27-O
| 4678. | +/- | 10. |
| 4764. | +/- | 111. |
| 4.14 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| -0.42 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.13 | +/- | 0. |
| 6.31 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.47 |
| -9999.00 |
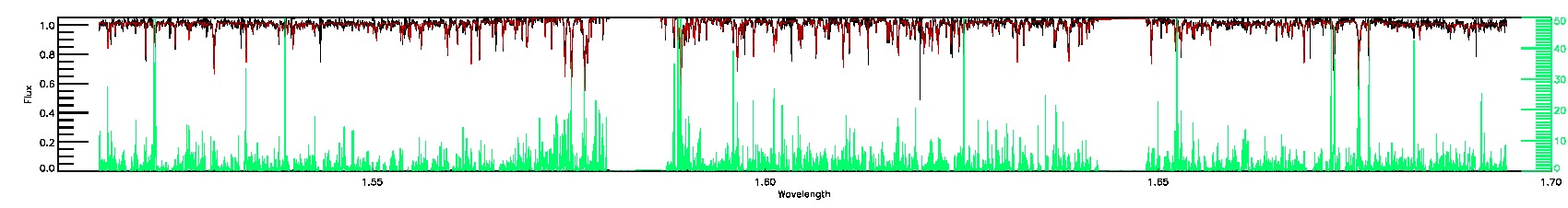
175-27-O
| 5226. | +/- | 15. |
| 5223. | +/- | 123. |
| 4.71 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.52 | +/- | 0. |
| 1.52 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.84 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.15 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.25 |
| -9999.00 |
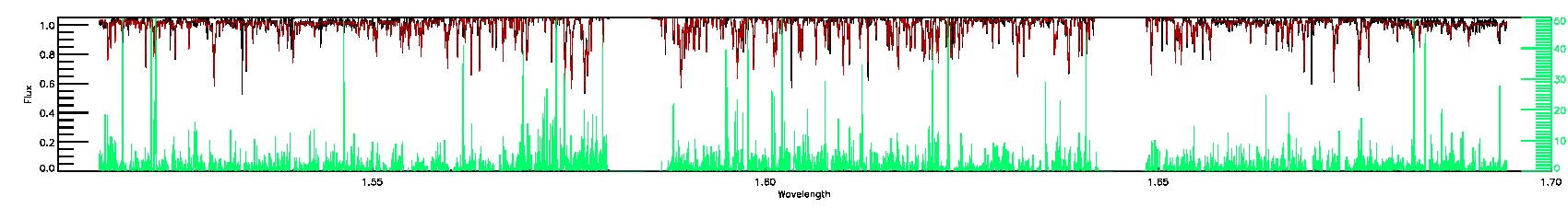
STAR_WARN,SN_WARN
175-27-O
| 5681. | +/- | 45. |
| 5631. | +/- | 155. |
| 4.56 | +/- | 0. |
| 4.48 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.64 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.98 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5845. | +/- | 45. |
| 5771. | +/- | 159. |
| 4.34 | +/- | 0. |
| 4.17 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.15 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 2.72 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.20 |
| -9999.00 |

175-27-O
| 5453. | +/- | 12. |
| 5427. | +/- | 118. |
| 4.67 | +/- | 0. |
| 4.59 | +/- | 0. |
| 1.85 | +/- | 0. |
| 1.85 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.68 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.83 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5717. | +/- | 57. |
| 5666. | +/- | 159. |
| 4.43 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.02 | +/- | 0. |
| 7.77 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.37 |
| -9999.00 |
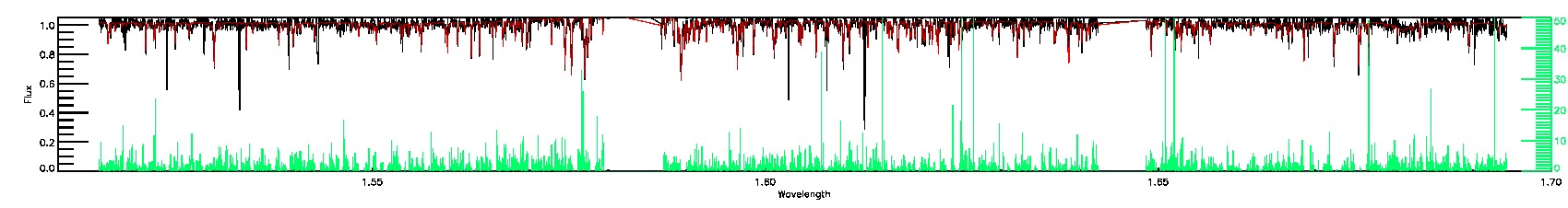
175-27-O
| 4275. | +/- | 1. |
| 4383. | +/- | 78. |
| 2.00 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.64 | +/- | 0. |
| 1.64 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -0.45 | +/- | 0. |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.23 | +/- | 0. |
| 3.86 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.75 |
| -9999.00 |
175-27-O
| 5481. | +/- | 13. |
| 5441. | +/- | 125. |
| 4.24 | +/- | 0. |
| 4.07 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.43 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.42 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 4979. | +/- | 62. |
| 5091. | +/- | 155. |
| 2.84 | +/- | 0. |
| 2.88 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -1.85 | +/- | 0. |
| -1.85 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.29 | +/- | 0. |
| 8.72 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.32 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.20 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5610. | +/- | 50. |
| 5577. | +/- | 155. |
| 4.47 | +/- | 0. |
| 4.48 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.61 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.25 |
| -9999.00 |
175-27-O
| 4505. | +/- | 4. |
| 4590. | +/- | 88. |
| 4.49 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.68 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
175-27-O
| 4504. | +/- | 11. |
| 4613. | +/- | 116. |
| 4.34 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 1.92 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.00 |
| -9999.00 |
175-27-O
| 4821. | +/- | 4. |
| 4866. | +/- | 85. |
| 2.67 | +/- | 0. |
| 2.43 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | 0. |
| -0.83 | +/- | 0. |
| -0.83 | +/- | -NaN |
| 0.56 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.73 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.87 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.03 |
| -9999.00 |
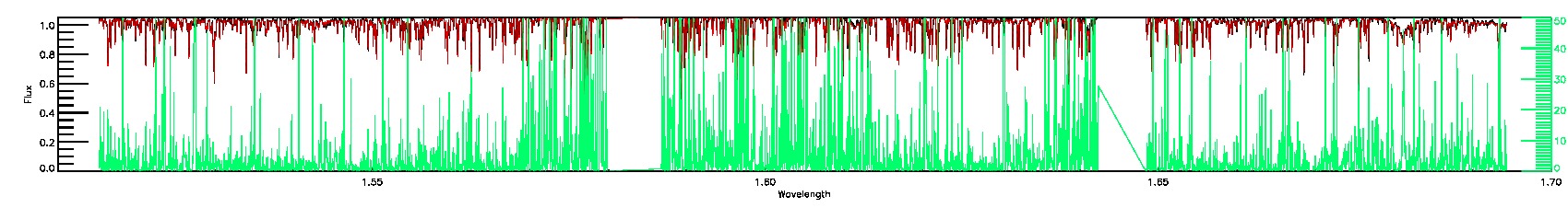
175-27-O
| 4878. | +/- | 5. |
| 4938. | +/- | 92. |
| 2.78 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.37 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.68 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |

175-27-O
| 4038. | +/- | 1. |
| 4135. | +/- | 70. |
| 1.67 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.55 | +/- | 0. |
| 1.55 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.32 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.37 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 6033. | +/- | 60. |
| 5951. | +/- | 175. |
| 4.33 | +/- | 0. |
| 4.26 | +/- | 0. |
| 0.83 | +/- | 0. |
| 0.83 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.08 | +/- | 0. |
| 2.63 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
175-27-O
| 4893. | +/- | 13. |
| 4952. | +/- | 115. |
| 3.18 | +/- | 0. |
| 3.05 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.68 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 1.00 |
| -9999.00 |
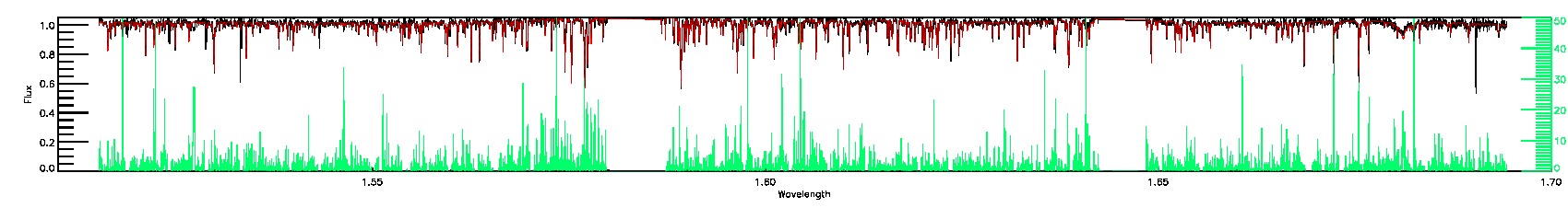
STAR_WARN,SN_WARN
175-27-O
| 4982. | +/- | 37. |
| 5040. | +/- | 137. |
| 2.76 | +/- | 0. |
| 2.42 | +/- | 0. |
| 1.73 | +/- | 0. |
| 1.73 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -0.59 | +/- | 0. |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.17 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.43 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 3448. | +/- | 4. |
| 3546. | +/- | 74. |
| 4.45 | +/- | 0. |
| 4.89 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.40 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 6.18 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.44 |
| -9999.00 |
175-27-O
| 4564. | +/- | 3. |
| 4632. | +/- | 83. |
| 3.18 | +/- | 0. |
| 2.95 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| 0.30 | +/- | 0. |
| 0.30 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.48 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.34 |
| -9999.00 |

BRIGHT_NEIGHBOR
175-27-O
| 5522. | +/- | 14. |
| 5492. | +/- | 120. |
| 4.62 | +/- | 0. |
| 4.58 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.54 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.23 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5380. | +/- | 48. |
| 5376. | +/- | 147. |
| 3.75 | +/- | 0. |
| 3.73 | +/- | 0. |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.58 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.30 |
| -9999.00 |

175-27-O
| 4968. | +/- | 6. |
| 4989. | +/- | 90. |
| 3.66 | +/- | 0. |
| 3.50 | +/- | 0. |
| 0.66 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.46 | +/- | 0. |
| 0.39 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.44 |
| -9999.00 |
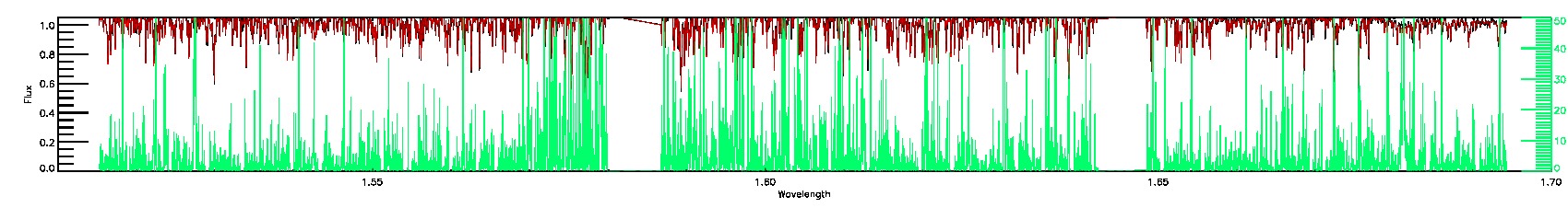
STAR_WARN,SN_WARN
175-27-O
| 4459. | +/- | 11. |
| 4559. | +/- | 112. |
| 4.33 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.50 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.37 |
| -9999.00 |
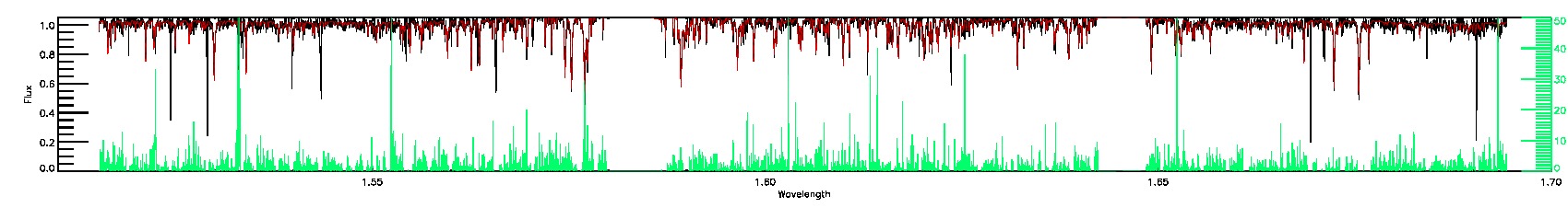
175-27-O
| 5774. | +/- | 27. |
| 5721. | +/- | 150. |
| 4.38 | +/- | 0. |
| 4.34 | +/- | 0. |
| 0.80 | +/- | 0. |
| 0.80 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.07 | +/- | 0. |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.29 |
| -9999.00 |

BRIGHT_NEIGHBOR
175-27-O
| 5596. | +/- | 13. |
| 5559. | +/- | 118. |
| 4.53 | +/- | 0. |
| 4.48 | +/- | 0. |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.65 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 4734. | +/- | 18. |
| 4805. | +/- | 120. |
| 4.46 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 1.80 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.40 |
| -9999.00 |

175-27-O
| 4657. | +/- | 5. |
| 4715. | +/- | 92. |
| 3.25 | +/- | 0. |
| 3.04 | +/- | 0. |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.50 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.99 |
| -9999.00 |

175-27-O
| 4422. | +/- | 3. |
| 4508. | +/- | 83. |
| 4.43 | +/- | 0. |
| 4.58 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.58 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.45 |
| -9999.00 |

SUSPECT_BROAD_LINES
175-27-O
| 6609. | +/- | 28. |
| 6453. | +/- | 170. |
| 4.45 | +/- | 0. |
| 4.20 | +/- | 0. |
| 2.62 | +/- | 0. |
| 2.62 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 31.67 | +/- | 0. |
| 1.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.00 |
| -9999.00 |
175-27-O
| 5260. | +/- | 15. |
| 5259. | +/- | 124. |
| 4.45 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.74 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 1.73 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| 0.08 |
| -9999.00 |

175-27-O
| 4445. | +/- | 3. |
| 4529. | +/- | 79. |
| 4.50 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.65 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 3.19 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.13 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5980. | +/- | 48. |
| 5895. | +/- | 168. |
| 4.43 | +/- | 0. |
| 4.27 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.09 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.32 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
175-27-O
| 4838. | +/- | 16. |
| 4885. | +/- | 115. |
| 3.41 | +/- | 0. |
| 3.24 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 2.86 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.67 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 4555. | +/- | 10. |
| 4634. | +/- | 108. |
| 4.50 | +/- | 0. |
| 4.61 | +/- | 0. |
| 0.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.50 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.12 |
| -9999.00 |

STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
175-27-O
| 5622. | +/- | 138. |
| -10000. | +/- | -NaN |
| 4.32 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.97 | +/- | 0. |
| 1.97 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.45 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 1.00 |
| -9999.00 |

STAR_BAD
175-27-O
| 5868. | +/- | 34. |
| -10000. | +/- | -NaN |
| 4.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.66 | +/- | 0. |
| 0.66 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.90 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.46 |
| -9999.00 |
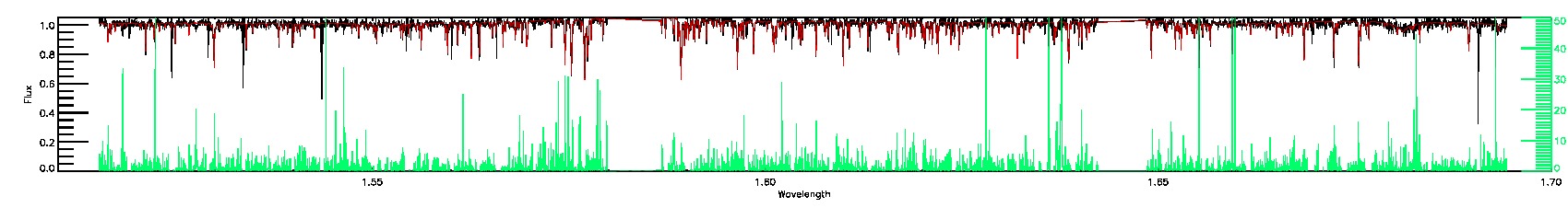
BRIGHT_NEIGHBOR
175-27-O
| 5704. | +/- | 25. |
| 5649. | +/- | 143. |
| 4.60 | +/- | 0. |
| 4.49 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.29 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.12 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.01 |
| -9999.00 |
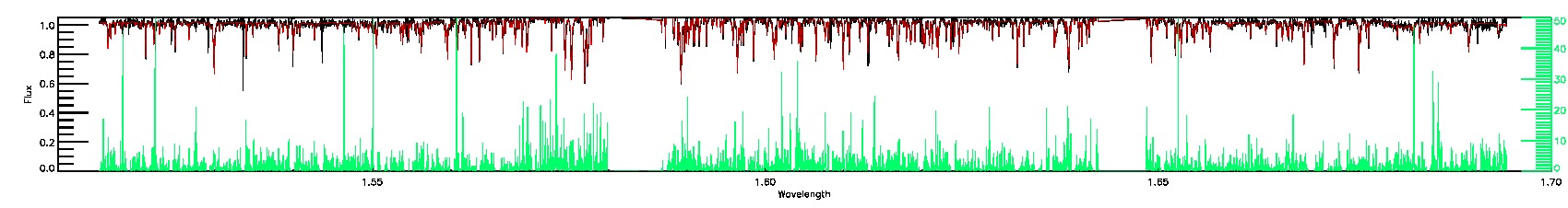
175-27-O
| 5262. | +/- | 24. |
| 5283. | +/- | 131. |
| 3.49 | +/- | 0. |
| 3.21 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.39 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -0.49 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 3.94 | +/- | 0. |
| 0.60 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.43 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 7258. | +/- | 13. |
| 7046. | +/- | 173. |
| 4.29 | +/- | 0. |
| 3.81 | +/- | 0. |
| 2.90 | +/- | 0. |
| 2.90 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 78.98 | +/- | 0. |
| 1.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.06 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5784. | +/- | 58. |
| 5736. | +/- | 162. |
| 4.26 | +/- | 0. |
| 4.29 | +/- | 0. |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 6.11 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
175-27-O
| 4616. | +/- | 7. |
| 4702. | +/- | 104. |
| 4.46 | +/- | 0. |
| 4.70 | +/- | 0. |
| 0.59 | +/- | 0. |
| 0.59 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 1.96 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.51 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5921. | +/- | 44. |
| 5855. | +/- | 166. |
| 4.47 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.98 | +/- | 0. |
| 0.98 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.13 | +/- | 0. |
| 2.03 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.67 |
| -9999.00 |
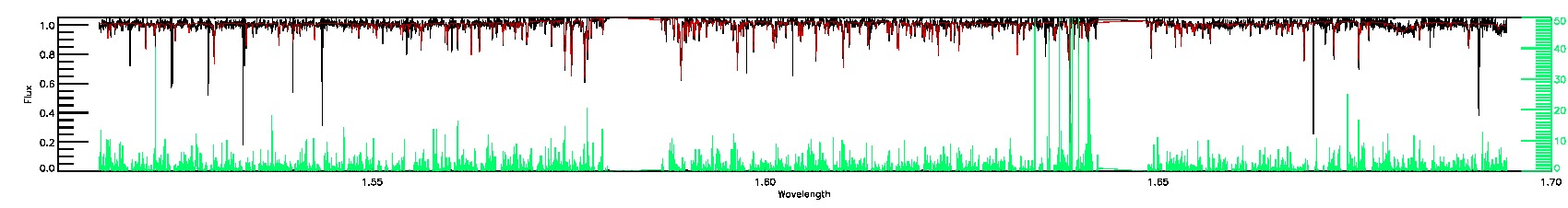
175-27-O
| 4124. | +/- | 1. |
| 4204. | +/- | 69. |
| 2.04 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.63 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.25 |
| -9999.00 |
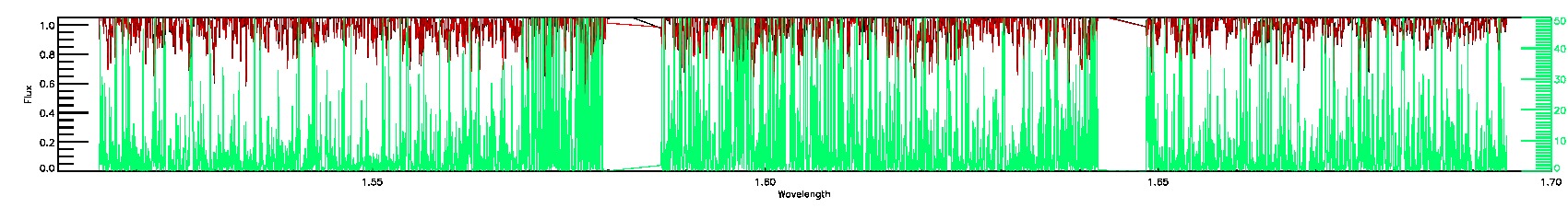
175-27-O
| 4936. | +/- | 6. |
| 4971. | +/- | 96. |
| 4.48 | +/- | 0. |
| 4.52 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.06 | +/- | 0. |
| 1.61 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.26 |
| -9999.00 |

175-27-O
| 4524. | +/- | 4. |
| 4624. | +/- | 87. |
| 2.49 | +/- | 0. |
| 2.31 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.61 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.22 |
| -9999.00 |
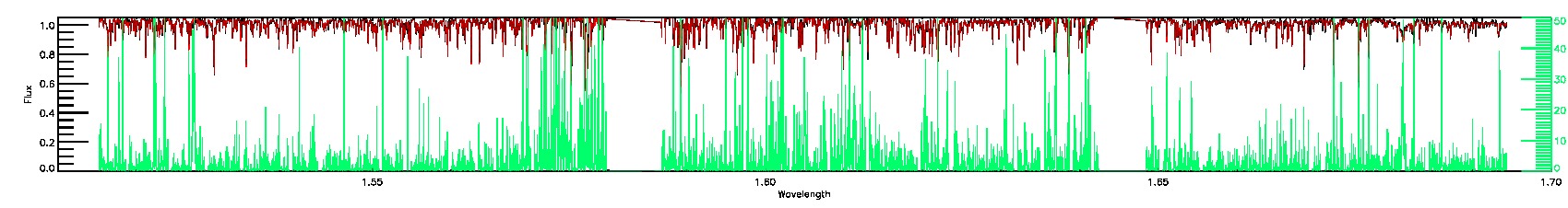
SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
175-27-O
| 7347. | +/- | 16. |
| -10000. | +/- | -NaN |
| 4.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.48 | +/- | 0. |
| 3.48 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 50.42 | +/- | 0. |
| 1.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.03 |
| -9999.00 |
STAR_WARN,SN_WARN
175-27-O
| 5445. | +/- | 28. |
| 5417. | +/- | 140. |
| 4.42 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.95 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.75 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 1.00 |
| -9999.00 |

175-27-O
| 4230. | +/- | 2. |
| 4338. | +/- | 90. |
| 1.91 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 3.87 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.90 |
| -9999.00 |
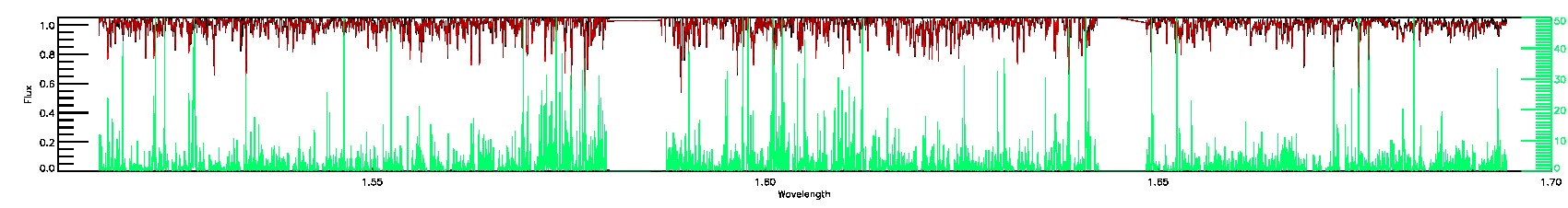
BRIGHT_NEIGHBOR
175-27-O
| 5872. | +/- | 28. |
| 5801. | +/- | 155. |
| 4.30 | +/- | 0. |
| 4.18 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 1.66 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 3884. | +/- | 5. |
| 3981. | +/- | 88. |
| 4.42 | +/- | 0. |
| 4.77 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
175-27-O
| 4738. | +/- | 5. |
| 4806. | +/- | 88. |
| 2.69 | +/- | 0. |
| 2.38 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.51 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.17 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.04 |
| -9999.00 |

STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
175-27-O
| 6796. | +/- | 68. |
| -10000. | +/- | -NaN |
| 4.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.11 | +/- | 0. |
| 3.11 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.72 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 1.39 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
175-27-O
| 4873. | +/- | 5. |
| 4931. | +/- | 91. |
| 2.77 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.53 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.53 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.03 |
| -9999.00 |

175-27-O
| 5418. | +/- | 19. |
| 5395. | +/- | 134. |
| 4.60 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 1.95 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.32 |
| -9999.00 |

175-27-O
| 5872. | +/- | 11. |
| 5804. | +/- | 119. |
| 4.40 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.82 | +/- | 0. |
| 0.82 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.28 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.01 | +/- | 0. |
| 1.60 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.17 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5429. | +/- | 36. |
| 5426. | +/- | 146. |
| 4.34 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.63 | +/- | 0. |
| 0.63 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.38 | +/- | 0. |
| 0.44 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 3.69 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.44 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.90 |
| -9999.00 |

175-27-O
| 5939. | +/- | 16. |
| 5856. | +/- | 134. |
| 4.48 | +/- | 0. |
| 4.31 | +/- | 0. |
| 0.79 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| 15.85 | +/- | 0. |
| 1.20 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.06 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 5003. | +/- | 9. |
| 5060. | +/- | 98. |
| 4.34 | +/- | 0. |
| 4.65 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -0.64 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.20 | +/- | 0. |
| 1.62 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.94 |
| -9999.00 |

175-27-O
| 4812. | +/- | 13. |
| 4876. | +/- | 113. |
| 3.07 | +/- | 0. |
| 2.92 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.20 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -0.28 | +/- | 0. |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.49 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.48 |
| -9999.00 |
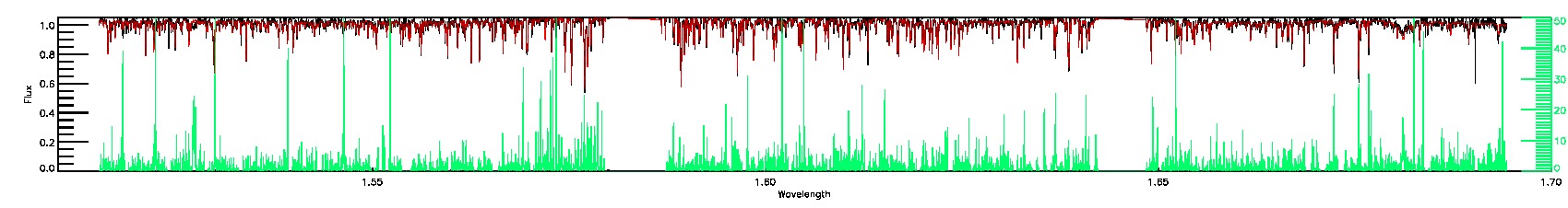
175-27-O
| 5708. | +/- | 12. |
| 5650. | +/- | 117. |
| 4.43 | +/- | 0. |
| 4.29 | +/- | 0. |
| 1.10 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.95 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.36 |
| -9999.00 |

175-27-O
| 5676. | +/- | 21. |
| 5642. | +/- | 143. |
| 4.59 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.35 | +/- | 0. |
| 0.35 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 1.52 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.35 |
| -9999.00 |

175-27-O
| 5716. | +/- | 28. |
| 5659. | +/- | 146. |
| 4.54 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.73 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 2.74 | +/- | 0. |
| 0.44 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.04 |
| -9999.00 |

175-27-O
| 4903. | +/- | 16. |
| 4947. | +/- | 118. |
| 3.40 | +/- | 0. |
| 3.24 | +/- | 0. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.07 | +/- | 0. |
| 0.49 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.20 |
| -9999.00 |
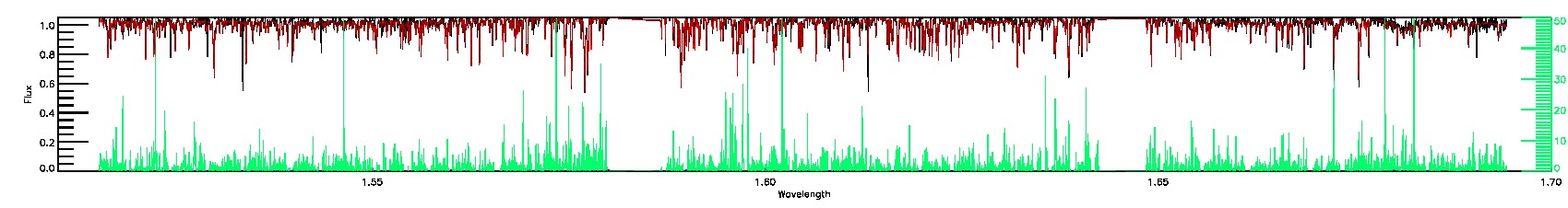
SUSPECT_BROAD_LINES
175-27-O
| 5294. | +/- | 16. |
| 5296. | +/- | 104. |
| 4.29 | +/- | 0. |
| 4.35 | +/- | 0. |
| 2.27 | +/- | 0. |
| 2.27 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.08 | +/- | 0. |
| 61.21 | +/- | 0. |
| 1.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.04 |
| -9999.00 |

175-27-O
| 4730. | +/- | 8. |
| 4792. | +/- | 104. |
| 4.53 | +/- | 0. |
| 4.63 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
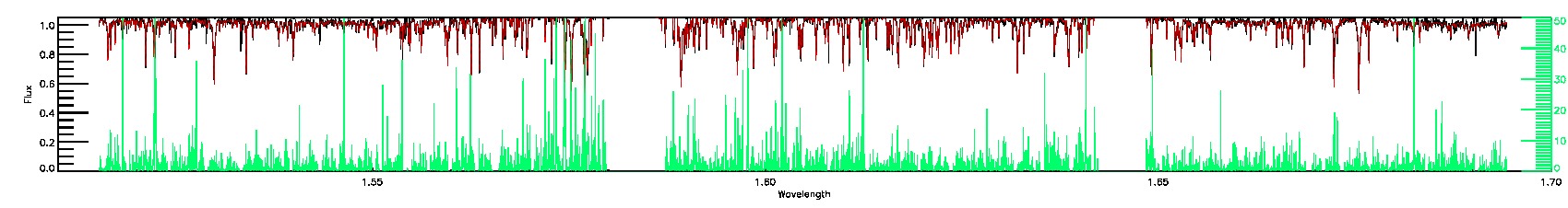
STAR_WARN,COLORTE_WARN,SN_WARN
175-27-O
| 4076. | +/- | 6. |
| 4169. | +/- | 93. |
| 4.45 | +/- | 0. |
| 4.72 | +/- | 0. |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 3.46 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |

175-27-O
| 5498. | +/- | 11. |
| 5471. | +/- | 108. |
| 4.39 | +/- | 0. |
| 4.36 | +/- | 0. |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.39 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
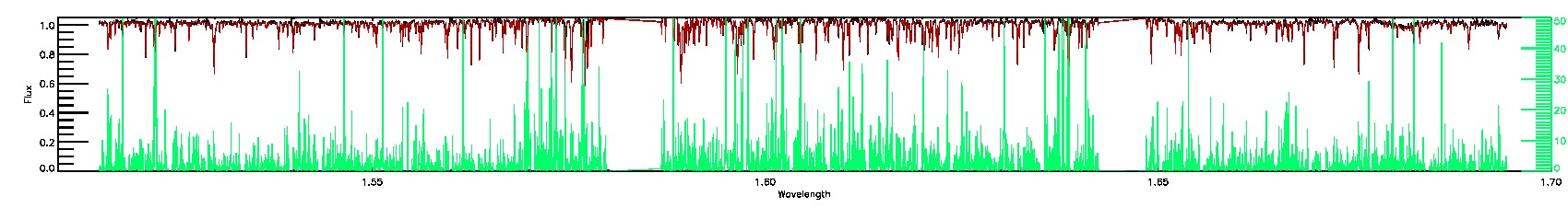
175-27-O
| 5477. | +/- | 13. |
| 5452. | +/- | 122. |
| 4.50 | +/- | 0. |
| 4.47 | +/- | 0. |
| 0.88 | +/- | 0. |
| 0.88 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.08 | +/- | 0. |
| -0.08 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.11 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.34 |
| -9999.00 |

175-27-O
| 4747. | +/- | 5. |
| 4813. | +/- | 92. |
| 2.78 | +/- | 0. |
| 2.44 | +/- | 0. |
| 1.45 | +/- | 0. |
| 1.45 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.21 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 6127. | +/- | 51. |
| 6031. | +/- | 174. |
| 4.56 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.92 | +/- | 0. |
| 0.92 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| 0.90 | +/- | 0. |
| 0.90 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.01 | +/- | 0. |
| 2.42 | +/- | 0. |
| 0.38 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.23 |
| -9999.00 |
175-27-O
| 5667. | +/- | 18. |
| 5618. | +/- | 131. |
| 4.29 | +/- | 0. |
| 4.20 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.19 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.06 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.01 | +/- | 0. |
| 3.04 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.81 |
| -9999.00 |
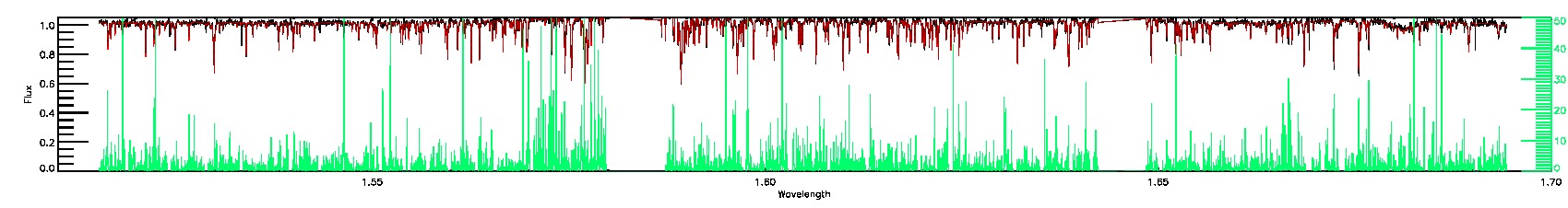
SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
175-27-O
| 7304. | +/- | 17. |
| -10000. | +/- | -NaN |
| 4.21 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.69 | +/- | 0. |
| 4.69 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 27.06 | +/- | 0. |
| 1.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.83 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.45 |
| -9999.00 |
175-27-O
| 4990. | +/- | 13. |
| 5041. | +/- | 115. |
| 2.87 | +/- | 0. |
| 2.51 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.76 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -0.46 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.87 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.49 |
| -9999.00 |
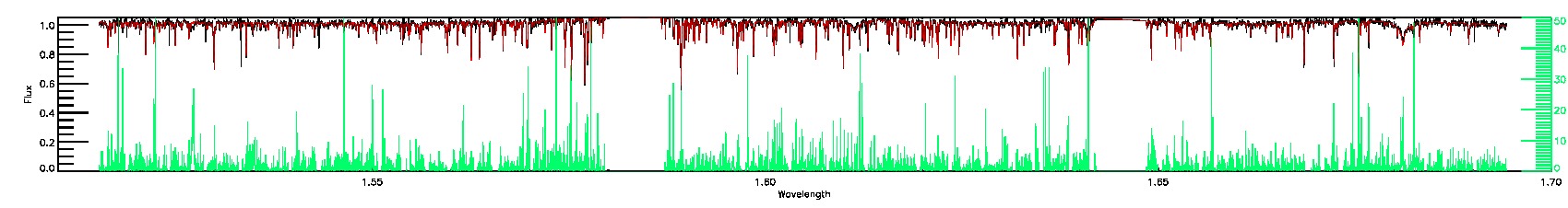
STAR_WARN,SN_WARN
175-27-O
| 5893. | +/- | 50. |
| 5833. | +/- | 168. |
| 4.06 | +/- | 0. |
| 4.07 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.31 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.66 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 6516. | +/- | 23. |
| 6382. | +/- | 148. |
| 4.98 | +/- | 0. |
| 4.86 | +/- | 0. |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.78 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
BRIGHT_NEIGHBOR
175-27-O
| 4934. | +/- | 14. |
| 4965. | +/- | 116. |
| 4.63 | +/- | 0. |
| 4.63 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.94 | +/- | 0. |
| 0.59 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.22 |
| -9999.00 |
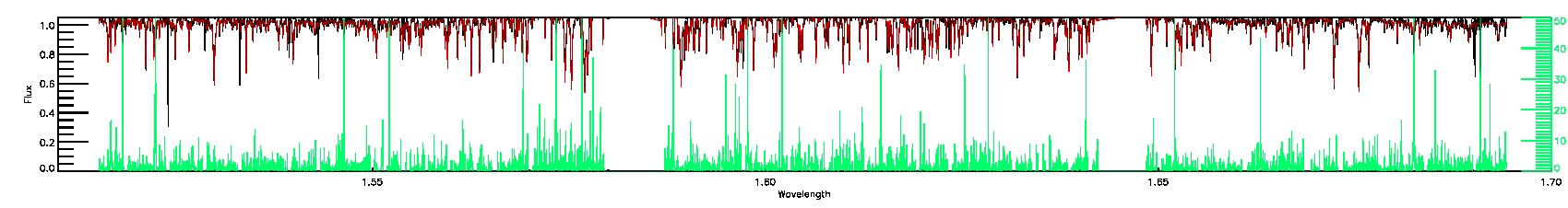
STAR_WARN,COLORTE_WARN
175-27-O
| 4141. | +/- | 4. |
| 4225. | +/- | 90. |
| 4.33 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 5.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.79 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
175-27-O
| 7455. | +/- | 11. |
| -10000. | +/- | -NaN |
| 4.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.58 | +/- | 0. |
| 4.58 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 29.80 | +/- | 0. |
| 1.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.24 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.81 |
| -9999.00 |
STAR_WARN,COLORTE_WARN
175-27-O
| 6488. | +/- | 53. |
| 6373. | +/- | 199. |
| 4.33 | +/- | 0. |
| 4.36 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -0.63 | +/- | 0. |
| -0.63 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.17 | +/- | 1. |
| 0.17 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.19 | +/- | 0. |
| 5.49 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.91 |
| -9999.00 |
175-27-O
| 5592. | +/- | 31. |
| 5556. | +/- | 144. |
| 4.58 | +/- | 0. |
| 4.54 | +/- | 0. |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.23 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.23 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.18 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.81 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5592. | +/- | 35. |
| 5547. | +/- | 146. |
| 4.04 | +/- | 0. |
| 3.92 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.04 | +/- | 0. |
| 2.72 | +/- | 0. |
| 0.43 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.01 |
| -9999.00 |
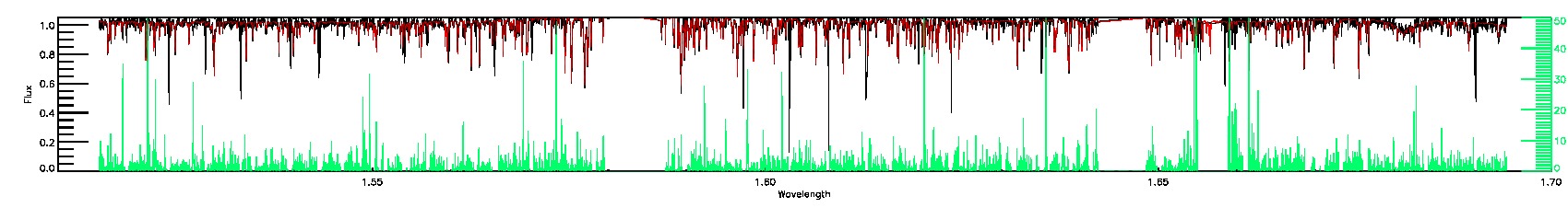
175-27-O
| 4707. | +/- | 4. |
| 4776. | +/- | 85. |
| 2.72 | +/- | 0. |
| 2.40 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 3.14 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.22 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 5269. | +/- | 30. |
| 5279. | +/- | 138. |
| 3.84 | +/- | 0. |
| 3.87 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 1.94 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.38 |
| -9999.00 |

175-27-O
| 4735. | +/- | 8. |
| 4803. | +/- | 104. |
| 3.25 | +/- | 0. |
| 3.10 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.13 | +/- | 0. |
| 3.27 | +/- | 0. |
| 0.51 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.14 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.82 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
175-27-O
| 6397. | +/- | 17. |
| 6272. | +/- | 141. |
| 4.40 | +/- | 0. |
| 4.25 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.85 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.04 |
| -9999.00 |
175-27-O
| 5624. | +/- | 21. |
| 5588. | +/- | 133. |
| 4.50 | +/- | 0. |
| 4.49 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.02 | +/- | 0. |
| 0.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.43 |
| -9999.00 |

175-27-O
| 5546. | +/- | 10. |
| 5511. | +/- | 112. |
| 4.43 | +/- | 0. |
| 4.35 | +/- | 0. |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 2.16 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.10 |
| -9999.00 |

STAR_WARN,SN_WARN
175-27-O
| 4531. | +/- | 9. |
| 4625. | +/- | 109. |
| 2.68 | +/- | 0. |
| 2.49 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.20 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.18 | +/- | 0. |
| 3.34 | +/- | 0. |
| 0.52 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |