STAR_BAD,SN_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 7131. | +/- | 129. |
| -10000. | +/- | -NaN |
| 4.70 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.50 | +/- | 0. |
| 0.50 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 | +/- | 2. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.81 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.19 |
| -9999.00 |
SMC6
| 5965. | +/- | 13. |
| 5893. | +/- | 125. |
| 4.12 | +/- | 0. |
| 4.09 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.22 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.95 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |

STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN
SMC6
| 3326. | +/- | 1. |
| -10000. | +/- | -NaN |
| 3.87 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.23 | +/- | 0. |
| 3.23 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.96 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 1.00 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4332. | +/- | 29. |
| 4489. | +/- | 128. |
| 1.32 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.32 | +/- | -NaN |
| -1.61 | +/- | 0. |
| -1.61 | +/- | 0. |
| -0.83 | +/- | 0. |
| -0.83 | +/- | -NaN |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 7.57 | +/- | 0. |
| 0.88 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.98 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.64 |
| -9999.00 |

SUSPECT_BROAD_LINES
SMC6
| 6466. | +/- | 22. |
| 6333. | +/- | 143. |
| 4.37 | +/- | 0. |
| 4.21 | +/- | 0. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 71.40 | +/- | 0. |
| 1.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.17 |
| -9999.00 |
LOW_SNR
STAR_WARN,SN_WARN
SMC6
| 4178. | +/- | 12. |
| 4318. | +/- | 115. |
| 1.44 | +/- | 0. |
| 1.42 | +/- | 0. |
| 2.33 | +/- | 0. |
| 2.33 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -1.21 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.39 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 5.99 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.66 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4150. | +/- | 10. |
| 4276. | +/- | 111. |
| 1.68 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.82 | +/- | 0. |
| 1.82 | +/- | -NaN |
| -0.87 | +/- | 0. |
| -0.87 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.91 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -2.50 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 3987. | +/- | 7. |
| 4130. | +/- | 107. |
| 0.99 | +/- | 0. |
| 1.02 | +/- | 0. |
| 2.36 | +/- | 0. |
| 2.36 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -1.29 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.44 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 6.28 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.32 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4106. | +/- | 13. |
| 4249. | +/- | 113. |
| 1.21 | +/- | 0. |
| 1.23 | +/- | 0. |
| 1.90 | +/- | 0. |
| 1.90 | +/- | -NaN |
| -1.28 | +/- | 0. |
| -1.28 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.25 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -2.27 |
| -9999.00 |

SMC6
| 4008. | +/- | 3. |
| 4138. | +/- | 96. |
| 1.03 | +/- | 0. |
| 1.01 | +/- | 0. |
| 2.04 | +/- | 0. |
| 2.04 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -0.98 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 5.23 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -0.81 |
| -9999.00 |

SMC6
| 4866. | +/- | 6. |
| 4912. | +/- | 91. |
| 3.44 | +/- | 0. |
| 3.28 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.95 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4272. | +/- | 16. |
| 4411. | +/- | 120. |
| 2.03 | +/- | 0. |
| 1.95 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -1.17 | +/- | 0. |
| -1.17 | +/- | 0. |
| -0.46 | +/- | 0. |
| -0.46 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 5.87 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.71 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 3769. | +/- | 16. |
| 3905. | +/- | 102. |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -1.13 | +/- | 0. |
| -1.13 | +/- | 0. |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.41 | +/- | 0. |
| 5.73 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.07 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4482. | +/- | 20. |
| 4607. | +/- | 123. |
| 1.98 | +/- | 0. |
| 1.86 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -0.86 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.19 | +/- | 0. |
| 4.89 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.95 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 5612. | +/- | 46. |
| 5573. | +/- | 153. |
| 4.53 | +/- | 0. |
| 4.49 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 3.62 | +/- | 0. |
| 0.56 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.23 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 3999. | +/- | 6. |
| 4132. | +/- | 103. |
| 1.26 | +/- | 0. |
| 1.24 | +/- | 0. |
| 2.13 | +/- | 0. |
| 2.13 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.46 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.03 |
| -9999.00 |

SMC6
| 3938. | +/- | 3. |
| 4067. | +/- | 96. |
| 0.88 | +/- | 0. |
| 0.86 | +/- | 0. |
| 2.07 | +/- | 0. |
| 2.07 | +/- | -NaN |
| -0.93 | +/- | 0. |
| -0.93 | +/- | 0. |
| -0.38 | +/- | 0. |
| -0.38 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.10 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.38 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.56 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4099. | +/- | 10. |
| 4252. | +/- | 111. |
| 0.80 | +/- | 0. |
| 0.85 | +/- | 0. |
| 2.36 | +/- | 0. |
| 2.36 | +/- | -NaN |
| -1.52 | +/- | 0. |
| -1.52 | +/- | 0. |
| -0.70 | +/- | 0. |
| -0.70 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 7.18 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.70 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.39 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4004. | +/- | 6. |
| 4141. | +/- | 103. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | 0. |
| 2.42 | +/- | 0. |
| 2.42 | +/- | -NaN |
| -1.13 | +/- | 0. |
| -1.13 | +/- | 0. |
| -0.46 | +/- | 0. |
| -0.46 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 5.74 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.16 |
| -9999.00 |

SMC6
| 3849. | +/- | 2. |
| 3983. | +/- | 93. |
| 0.56 | +/- | 0. |
| 0.55 | +/- | 0. |
| 2.26 | +/- | 0. |
| 2.26 | +/- | -NaN |
| -1.07 | +/- | 0. |
| -1.07 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.47 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.52 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -2.02 |
| -9999.00 |
SMC6
| 3945. | +/- | 3. |
| 4120. | +/- | 105. |
| 0.03 | +/- | 0. |
| 0.15 | +/- | 0. |
| 2.84 | +/- | 0. |
| 2.84 | +/- | -NaN |
| -2.04 | +/- | 0. |
| -2.04 | +/- | 0. |
| -0.69 | +/- | 0. |
| -0.69 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.11 | +/- | 0. |
| 9.75 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.70 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.35 |
| -9999.00 |
STAR_BAD
SMC6
| 3739. | +/- | 1. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.27 | +/- | 0. |
| 2.27 | +/- | -NaN |
| -0.84 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.84 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| 0.89 |
| -9999.00 |
SUSPECT_BROAD_LINES
SMC6
| 6524. | +/- | 17. |
| 6383. | +/- | 145. |
| 4.37 | +/- | 0. |
| 4.18 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 18.23 | +/- | 0. |
| 1.26 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4473. | +/- | 14. |
| 4600. | +/- | 116. |
| 1.92 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.93 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -0.90 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.37 | +/- | 0. |
| 0.37 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.99 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.35 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.20 |
| -9999.00 |

STAR_WARN,COLORTE_WARN
SMC6
| 6445. | +/- | 16. |
| 6308. | +/- | 140. |
| 4.66 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.23 | +/- | -NaN |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.02 | +/- | 0. |
| 6.79 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
SMC6
| 4253. | +/- | 6. |
| 4379. | +/- | 104. |
| 1.63 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.99 | +/- | 0. |
| 1.99 | +/- | -NaN |
| -0.87 | +/- | 0. |
| -0.87 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.91 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.77 |
| -9999.00 |

SMC6
| 3896. | +/- | 3. |
| 4027. | +/- | 93. |
| 0.75 | +/- | 0. |
| 0.73 | +/- | 0. |
| 2.12 | +/- | 0. |
| 2.12 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -0.98 | +/- | 0. |
| -0.38 | +/- | 0. |
| -0.38 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.00 | +/- | 0. |
| 5.25 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.93 |
| -9999.00 |
SMC6
| 4015. | +/- | 3. |
| 4149. | +/- | 97. |
| 1.08 | +/- | 0. |
| 1.07 | +/- | 0. |
| 2.28 | +/- | 0. |
| 2.28 | +/- | -NaN |
| -1.07 | +/- | 0. |
| -1.07 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.53 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.26 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4197. | +/- | 15. |
| 4355. | +/- | 121. |
| 0.95 | +/- | 0. |
| 1.02 | +/- | 0. |
| 2.34 | +/- | 0. |
| 2.34 | +/- | -NaN |
| -1.64 | +/- | 0. |
| -1.63 | +/- | 0. |
| -0.66 | +/- | 0. |
| -0.66 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 7.70 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.73 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.49 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4116. | +/- | 7. |
| 4247. | +/- | 106. |
| 1.37 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.97 | +/- | 0. |
| 1.97 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -0.98 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.25 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4071. | +/- | 10. |
| 4204. | +/- | 108. |
| 1.39 | +/- | 0. |
| 1.36 | +/- | 0. |
| 2.16 | +/- | 0. |
| 2.16 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.47 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.05 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4170. | +/- | 8. |
| 4295. | +/- | 109. |
| 1.15 | +/- | 0. |
| 1.11 | +/- | 0. |
| 2.53 | +/- | 0. |
| 2.53 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -0.86 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.88 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.83 |
| -9999.00 |

SMC6
| 4036. | +/- | 5. |
| 4190. | +/- | 107. |
| 0.91 | +/- | 0. |
| 0.97 | +/- | 0. |
| 2.15 | +/- | 0. |
| 2.15 | +/- | -NaN |
| -1.54 | +/- | 0. |
| -1.54 | +/- | 0. |
| -0.46 | +/- | 0. |
| -0.46 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.17 | +/- | 0. |
| 7.27 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.87 |
| -9999.00 |

SMC6
| 4076. | +/- | 6. |
| 4210. | +/- | 102. |
| 1.07 | +/- | 0. |
| 1.06 | +/- | 0. |
| 2.01 | +/- | 0. |
| 2.01 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -1.07 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 5.55 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| 0.01 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4207. | +/- | 16. |
| 4358. | +/- | 120. |
| 1.48 | +/- | 0. |
| 1.49 | +/- | 0. |
| 2.28 | +/- | 0. |
| 2.28 | +/- | -NaN |
| -1.45 | +/- | 0. |
| -1.45 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.44 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.91 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -2.14 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4196. | +/- | 10. |
| 4324. | +/- | 113. |
| 1.67 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.85 | +/- | 0. |
| 1.85 | +/- | -NaN |
| -0.92 | +/- | 0. |
| -0.92 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.23 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 5.08 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.91 |
| -9999.00 |

SMC6
| 6204. | +/- | 19. |
| 6105. | +/- | 134. |
| 4.42 | +/- | 0. |
| 4.34 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.52 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.13 |
| -9999.00 |
STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN,COLORTE_WARN
SMC6
| 3101. | +/- | 1. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.44 | +/- | 0. |
| 2.44 | +/- | -NaN |
| -1.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.27 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.24 |
| -9999.00 |
BRIGHT_NEIGHBOR,VERY_BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4203. | +/- | 11. |
| 4339. | +/- | 115. |
| 1.78 | +/- | 0. |
| 1.70 | +/- | 0. |
| 1.72 | +/- | 0. |
| 1.72 | +/- | -NaN |
| -1.10 | +/- | 0. |
| -1.10 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.64 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.88 |
| -9999.00 |

SMC6
| 3852. | +/- | 3. |
| 3987. | +/- | 94. |
| 0.71 | +/- | 0. |
| 0.71 | +/- | 0. |
| 2.42 | +/- | 0. |
| 2.42 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -1.08 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.50 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.57 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.49 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.60 |
| -9999.00 |
LOW_SNR
STAR_WARN,SN_WARN
SMC6
| 4197. | +/- | 14. |
| 4333. | +/- | 117. |
| 1.56 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.87 | +/- | 0. |
| 1.87 | +/- | -NaN |
| -1.12 | +/- | 0. |
| -1.12 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.47 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.70 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.80 |
| -9999.00 |

SMC6
| 3979. | +/- | 3. |
| 4110. | +/- | 98. |
| 1.22 | +/- | 0. |
| 1.20 | +/- | 0. |
| 2.24 | +/- | 0. |
| 2.24 | +/- | -NaN |
| -0.99 | +/- | 0. |
| -0.99 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.27 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.27 |
| -9999.00 |
SMC6
| 3942. | +/- | 4. |
| 4067. | +/- | 98. |
| 1.04 | +/- | 0. |
| 1.00 | +/- | 0. |
| 2.04 | +/- | 0. |
| 2.04 | +/- | -NaN |
| -0.87 | +/- | 0. |
| -0.87 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.06 | +/- | 0. |
| 4.93 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.80 |
| -9999.00 |
LOW_SNR
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 4323. | +/- | 18. |
| 4450. | +/- | 119. |
| 1.85 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.61 | +/- | 0. |
| 1.61 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -0.90 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 5.00 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.86 |
| -9999.00 |

SMC6
| 3920. | +/- | 3. |
| 4046. | +/- | 95. |
| 1.00 | +/- | 0. |
| 0.97 | +/- | 0. |
| 2.14 | +/- | 0. |
| 2.14 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -0.88 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 4.95 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -1.79 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4221. | +/- | 17. |
| 4362. | +/- | 120. |
| 1.85 | +/- | 0. |
| 1.79 | +/- | 0. |
| 3.24 | +/- | 0. |
| 3.24 | +/- | -NaN |
| -1.24 | +/- | 0. |
| -1.24 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.09 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.22 |
| -9999.00 |

SMC6
| 5252. | +/- | 13. |
| 5267. | +/- | 115. |
| 3.77 | +/- | 0. |
| 3.80 | +/- | 0. |
| 1.46 | +/- | 0. |
| 1.46 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 1.66 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.27 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4161. | +/- | 11. |
| 4295. | +/- | 111. |
| 1.38 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.78 | +/- | 0. |
| 1.78 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -1.08 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.56 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.35 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.76 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4233. | +/- | 18. |
| 4401. | +/- | 125. |
| 0.92 | +/- | 0. |
| 1.02 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -1.87 | +/- | 0. |
| -1.87 | +/- | 0. |
| -0.61 | +/- | 0. |
| -0.61 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.22 | +/- | 0. |
| 8.81 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.81 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.22 |
| -9999.00 |

SMC6
| 3760. | +/- | 2. |
| 3894. | +/- | 87. |
| 0.38 | +/- | 0. |
| 0.37 | +/- | 0. |
| 2.59 | +/- | 0. |
| 2.59 | +/- | -NaN |
| -1.07 | +/- | 0. |
| -1.07 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.39 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.53 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.98 |
| -9999.00 |
SMC6
| 3920. | +/- | 4. |
| 4054. | +/- | 97. |
| 0.80 | +/- | 0. |
| 0.79 | +/- | 0. |
| 2.10 | +/- | 0. |
| 2.10 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.53 | +/- | 0. |
| -0.53 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.47 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.01 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4250. | +/- | 7. |
| 4370. | +/- | 108. |
| 1.87 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.35 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -0.73 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.53 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.78 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4353. | +/- | 13. |
| 4473. | +/- | 113. |
| 2.00 | +/- | 0. |
| 1.86 | +/- | 0. |
| 1.53 | +/- | 0. |
| 1.53 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -0.73 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.54 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.30 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4190. | +/- | 8. |
| 4312. | +/- | 107. |
| 1.50 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.68 | +/- | -NaN |
| -0.80 | +/- | 0. |
| -0.80 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.28 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 4.73 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.74 |
| -9999.00 |

STAR_BAD
SMC6
| 3628. | +/- | 1. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.59 | +/- | 0. |
| 2.59 | +/- | -NaN |
| -1.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.33 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.52 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4178. | +/- | 9. |
| 4298. | +/- | 109. |
| 2.07 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.73 | +/- | 0. |
| 1.73 | +/- | -NaN |
| -0.72 | +/- | 0. |
| -0.72 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.51 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.73 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.59 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR
SMC6
| 3978. | +/- | 3. |
| 4111. | +/- | 100. |
| 1.21 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.77 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 5.47 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -0.42 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,COLORTE_WARN,SN_WARN
SMC6
| 5884. | +/- | 107. |
| -10000. | +/- | -NaN |
| 4.35 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.33 | +/- | 0. |
| 0.33 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.75 | +/- | 0. |
| 0.83 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.20 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR
SMC6
| 4219. | +/- | 6. |
| 4364. | +/- | 105. |
| 1.19 | +/- | 0. |
| 1.22 | +/- | 0. |
| 2.07 | +/- | 0. |
| 2.07 | +/- | -NaN |
| -1.33 | +/- | 0. |
| -1.33 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.50 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.06 | +/- | 0. |
| 6.44 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.51 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.31 |
| -9999.00 |

SMC6
| 6114. | +/- | 38. |
| 6031. | +/- | 162. |
| 4.43 | +/- | 0. |
| 4.43 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.14 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 3.35 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.82 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4018. | +/- | 5. |
| 4150. | +/- | 102. |
| 1.16 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -1.03 | +/- | 0. |
| -0.37 | +/- | 0. |
| -0.37 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.40 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.35 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.93 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4206. | +/- | 17. |
| 4358. | +/- | 119. |
| 1.20 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.95 | +/- | 0. |
| 1.95 | +/- | -NaN |
| -1.47 | +/- | 0. |
| -1.47 | +/- | 0. |
| -0.98 | +/- | 0. |
| -0.98 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 7.00 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.99 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.47 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4182. | +/- | 10. |
| 4307. | +/- | 111. |
| 1.42 | +/- | 0. |
| 1.35 | +/- | 0. |
| 1.79 | +/- | 0. |
| 1.79 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -0.86 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.28 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.88 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.18 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4174. | +/- | 11. |
| 4309. | +/- | 113. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | 0. |
| 2.49 | +/- | 0. |
| 2.49 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -1.08 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.39 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.07 | +/- | 0. |
| 5.58 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -2.01 |
| -9999.00 |

SMC6
| 3922. | +/- | 2. |
| 4048. | +/- | 92. |
| 0.87 | +/- | 0. |
| 0.84 | +/- | 0. |
| 2.31 | +/- | 0. |
| 2.31 | +/- | -NaN |
| -0.89 | +/- | 0. |
| -0.89 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.98 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.35 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.86 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4154. | +/- | 6. |
| 4279. | +/- | 105. |
| 1.43 | +/- | 0. |
| 1.37 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.81 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -0.88 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 4.94 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.84 |
| -9999.00 |
SMC6
| 3940. | +/- | 3. |
| 4074. | +/- | 97. |
| 0.89 | +/- | 0. |
| 0.89 | +/- | 0. |
| 2.09 | +/- | 0. |
| 2.09 | +/- | -NaN |
| -1.07 | +/- | 0. |
| -1.07 | +/- | 0. |
| -0.45 | +/- | 0. |
| -0.45 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.53 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.15 |
| -9999.00 |
SMC6
| 5410. | +/- | 20. |
| 5393. | +/- | 132. |
| 4.43 | +/- | 0. |
| 4.41 | +/- | 0. |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.00 | +/- | 0. |
| 3.47 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.10 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4017. | +/- | 9. |
| 4149. | +/- | 106. |
| 1.03 | +/- | 0. |
| 1.02 | +/- | 0. |
| 2.11 | +/- | 0. |
| 2.11 | +/- | -NaN |
| -1.01 | +/- | 0. |
| -1.01 | +/- | 0. |
| -0.60 | +/- | 0. |
| -0.60 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.15 | +/- | 0. |
| 5.35 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.54 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.12 |
| -9999.00 |

SMC6
| 6758. | +/- | 15. |
| 6591. | +/- | 157. |
| 4.45 | +/- | 0. |
| 4.24 | +/- | 0. |
| 0.93 | +/- | 0. |
| 0.93 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.17 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 7.43 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.46 |
| -9999.00 |
VERY_BRIGHT_NEIGHBOR
STAR_BAD
SMC6
| 3743. | +/- | 1. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.28 | +/- | 0. |
| 2.28 | +/- | -NaN |
| -0.95 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.16 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.43 |
| -9999.00 |
LOW_SNR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
SMC6
| 5050. | +/- | 110. |
| -10000. | +/- | -NaN |
| 2.76 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.37 | +/- | 0. |
| 1.37 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.89 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.22 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.13 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 4226. | +/- | 15. |
| 4367. | +/- | 120. |
| 1.12 | +/- | 0. |
| 1.13 | +/- | 0. |
| 2.34 | +/- | 0. |
| 2.34 | +/- | -NaN |
| -1.24 | +/- | 0. |
| -1.24 | +/- | 0. |
| -0.63 | +/- | 0. |
| -0.63 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| 6.10 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.60 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.54 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4142. | +/- | 8. |
| 4283. | +/- | 111. |
| 1.35 | +/- | 0. |
| 1.34 | +/- | 0. |
| 1.91 | +/- | 0. |
| 1.91 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -1.22 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 6.05 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.90 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.60 |
| -9999.00 |

STAR_BAD
SMC6
| 3606. | +/- | 6. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.86 | +/- | 0. |
| 1.86 | +/- | -NaN |
| -1.53 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.25 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.30 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.52 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
SMC6
| 4234. | +/- | 17. |
| -10000. | +/- | -NaN |
| 1.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.68 | +/- | 0. |
| 1.68 | +/- | -NaN |
| -1.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.71 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.06 |
| -9999.00 |

STAR_WARN,CHI2_WARN
SMC6
| 3208. | +/- | 1. |
| 3359. | +/- | 68. |
| 0.57 | +/- | 0. |
| 0.61 | +/- | 0. |
| 2.93 | +/- | 0. |
| 2.93 | +/- | -NaN |
| -1.47 | +/- | 0. |
| -1.47 | +/- | 0. |
| 0.64 | +/- | 0. |
| 0.64 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 6.97 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.63 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.45 |
| -9999.00 |
SMC6
| 4081. | +/- | 3. |
| 4204. | +/- | 95. |
| 1.39 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.89 | +/- | 0. |
| 1.89 | +/- | -NaN |
| -0.81 | +/- | 0. |
| -0.81 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.82 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4171. | +/- | 9. |
| 4290. | +/- | 107. |
| 1.65 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.75 | +/- | 0. |
| 1.75 | +/- | -NaN |
| -0.72 | +/- | 0. |
| -0.72 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.10 | +/- | 0. |
| 4.50 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -0.29 |
| -9999.00 |

SMC6
| 3681. | +/- | 1. |
| 3817. | +/- | 74. |
| -0.22 | +/- | 0. |
| -0.22 | +/- | 0. |
| 2.59 | +/- | 0. |
| 2.59 | +/- | -NaN |
| -1.12 | +/- | 0. |
| -1.12 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.69 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.97 |
| -9999.00 |
STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN
SMC6
| 3049. | +/- | 1. |
| -10000. | +/- | -NaN |
| 0.36 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.37 | +/- | 0. |
| 3.37 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.84 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.17 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.33 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4165. | +/- | 8. |
| 4300. | +/- | 108. |
| 1.38 | +/- | 0. |
| 1.36 | +/- | 0. |
| 2.05 | +/- | 0. |
| 2.05 | +/- | -NaN |
| -1.09 | +/- | 0. |
| -1.09 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.61 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.54 |
| -9999.00 |

SMC6
| 4031. | +/- | 4. |
| 4158. | +/- | 97. |
| 1.14 | +/- | 0. |
| 1.11 | +/- | 0. |
| 1.88 | +/- | 0. |
| 1.88 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -0.90 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.01 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.89 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4068. | +/- | 8. |
| 4197. | +/- | 108. |
| 1.33 | +/- | 0. |
| 1.30 | +/- | 0. |
| 2.08 | +/- | 0. |
| 2.08 | +/- | -NaN |
| -0.95 | +/- | 0. |
| -0.95 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.16 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.85 |
| -9999.00 |

SMC6
| 4186. | +/- | 8. |
| 4356. | +/- | 118. |
| 0.91 | +/- | 0. |
| 1.02 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -1.92 | +/- | 0. |
| -1.92 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.44 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| 0.41 | +/- | 0. |
| 0.38 | +/- | 0. |
| 9.08 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.54 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4042. | +/- | 9. |
| 4176. | +/- | 107. |
| 1.20 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.63 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.48 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -1.05 |
| -9999.00 |

SMC6
| 4095. | +/- | 6. |
| 4226. | +/- | 103. |
| 1.29 | +/- | 0. |
| 1.27 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -0.98 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.25 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -1.91 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4092. | +/- | 15. |
| 4218. | +/- | 109. |
| 1.16 | +/- | 0. |
| 1.13 | +/- | 0. |
| 1.30 | +/- | 0. |
| 1.30 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -0.88 | +/- | 0. |
| -0.18 | +/- | 0. |
| -0.18 | +/- | -NaN |
| 0.78 | +/- | 0. |
| 0.78 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.33 | +/- | 0. |
| 4.95 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.20 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.28 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4116. | +/- | 10. |
| 4257. | +/- | 111. |
| 1.17 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.95 | +/- | 0. |
| 1.95 | +/- | -NaN |
| -1.24 | +/- | 0. |
| -1.24 | +/- | 0. |
| -0.58 | +/- | 0. |
| -0.58 | +/- | -NaN |
| 0.25 | +/- | 0. |
| 0.25 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 6.12 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.54 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -2.19 |
| -9999.00 |
SMC6
| 6135. | +/- | 21. |
| 6049. | +/- | 136. |
| 4.36 | +/- | 0. |
| 4.36 | +/- | 0. |
| 1.25 | +/- | 0. |
| 1.25 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.38 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.03 |
| -9999.00 |
BRIGHT_NEIGHBOR,VERY_BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4444. | +/- | 9. |
| 4545. | +/- | 108. |
| 3.99 | +/- | 0. |
| 4.26 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | -NaN |
| -0.30 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 12.67 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.54 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4076. | +/- | 18. |
| 4227. | +/- | 117. |
| 0.75 | +/- | 0. |
| 0.79 | +/- | 0. |
| 2.14 | +/- | 0. |
| 2.14 | +/- | -NaN |
| -1.47 | +/- | 0. |
| -1.47 | +/- | 0. |
| -0.90 | +/- | 0. |
| -0.90 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -0.26 | +/- | 0. |
| 6.98 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.77 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.34 |
| -9999.00 |
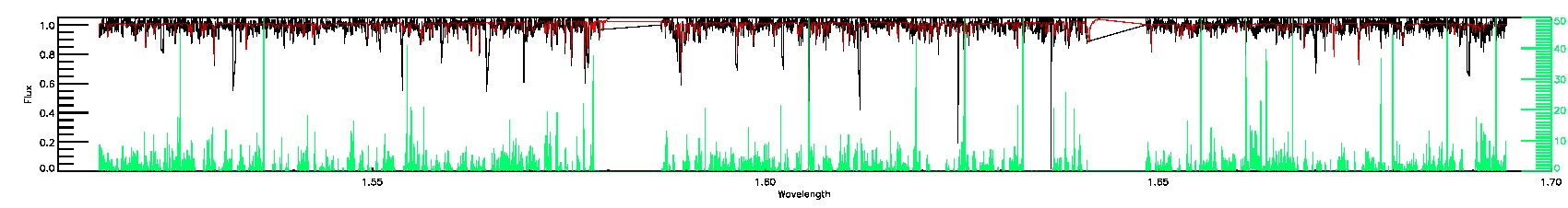
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4707. | +/- | 30. |
| 4811. | +/- | 131. |
| 2.15 | +/- | 0. |
| 2.04 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -0.91 | +/- | 0. |
| -0.91 | +/- | 0. |
| -0.69 | +/- | 0. |
| -0.69 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.36 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.18 | +/- | 0. |
| 5.04 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.86 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -2.17 |
| -9999.00 |

SMC6
| 5506. | +/- | 15. |
| 5485. | +/- | 121. |
| 3.99 | +/- | 0. |
| 4.01 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 4.13 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.19 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4313. | +/- | 14. |
| 4434. | +/- | 116. |
| 1.59 | +/- | 0. |
| 1.49 | +/- | 0. |
| 2.22 | +/- | 0. |
| 2.22 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -0.75 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.13 | +/- | 0. |
| 4.59 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -1.16 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 3832. | +/- | 11. |
| 3948. | +/- | 93. |
| 4.39 | +/- | 0. |
| 4.93 | +/- | 0. |
| 0.89 | +/- | 0. |
| 0.89 | +/- | -NaN |
| -0.65 | +/- | 0. |
| -0.64 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.37 | +/- | 0. |
| -0.37 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 7.30 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.37 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4081. | +/- | 9. |
| 4220. | +/- | 109. |
| 0.97 | +/- | 0. |
| 0.98 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.83 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -1.18 | +/- | 0. |
| -0.59 | +/- | 0. |
| -0.59 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.89 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.57 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -2.18 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4140. | +/- | 7. |
| 4283. | +/- | 110. |
| 1.12 | +/- | 0. |
| 1.14 | +/- | 0. |
| 2.20 | +/- | 0. |
| 2.20 | +/- | -NaN |
| -1.27 | +/- | 0. |
| -1.27 | +/- | 0. |
| -0.65 | +/- | 0. |
| -0.65 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.21 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.62 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.42 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 3989. | +/- | 5. |
| 4120. | +/- | 103. |
| 1.25 | +/- | 0. |
| 1.23 | +/- | 0. |
| 2.26 | +/- | 0. |
| 2.26 | +/- | -NaN |
| -1.01 | +/- | 0. |
| -1.01 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.33 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.81 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -1.93 |
| -9999.00 |

SMC6
| 6704. | +/- | 16. |
| 6543. | +/- | 154. |
| 4.65 | +/- | 0. |
| 4.45 | +/- | 0. |
| 1.67 | +/- | 0. |
| 1.67 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.37 | +/- | 0. |
| 0.53 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4138. | +/- | 10. |
| 4279. | +/- | 113. |
| 1.14 | +/- | 0. |
| 1.15 | +/- | 0. |
| 1.70 | +/- | 0. |
| 1.70 | +/- | -NaN |
| -1.23 | +/- | 0. |
| -1.23 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.15 | +/- | 0. |
| 6.07 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.14 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4001. | +/- | 7. |
| 4141. | +/- | 108. |
| 0.58 | +/- | 0. |
| 0.59 | +/- | 0. |
| 2.33 | +/- | 0. |
| 2.33 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -1.21 | +/- | 0. |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.06 | +/- | 0. |
| 5.99 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.14 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4161. | +/- | 20. |
| 4300. | +/- | 118. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | 0. |
| 2.48 | +/- | 0. |
| 2.48 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -1.18 | +/- | 0. |
| -0.52 | +/- | 0. |
| -0.52 | +/- | -NaN |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -0.11 | +/- | 0. |
| 5.89 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.44 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4222. | +/- | 14. |
| 4353. | +/- | 116. |
| 1.57 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.94 | +/- | 0. |
| 1.94 | +/- | -NaN |
| -1.01 | +/- | 0. |
| -1.01 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.23 | +/- | -NaN |
| -0.18 | +/- | 0. |
| -0.18 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.35 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.24 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4183. | +/- | 12. |
| 4330. | +/- | 116. |
| 1.28 | +/- | 0. |
| 1.31 | +/- | 0. |
| 2.45 | +/- | 0. |
| 2.45 | +/- | -NaN |
| -1.36 | +/- | 0. |
| -1.36 | +/- | 0. |
| -0.72 | +/- | 0. |
| -0.72 | +/- | -NaN |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.57 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.65 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.28 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 3966. | +/- | 8. |
| 4058. | +/- | 95. |
| 4.46 | +/- | 0. |
| 4.75 | +/- | 0. |
| 0.64 | +/- | 0. |
| 0.64 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.06 | +/- | 0. |
| -0.07 | +/- | 0. |
| 6.01 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.36 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4212. | +/- | 12. |
| 4345. | +/- | 116. |
| 1.31 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.91 | +/- | 0. |
| 1.91 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.11 | +/- | 0. |
| 5.46 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.25 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4239. | +/- | 10. |
| 4359. | +/- | 112. |
| 1.60 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.75 | +/- | 0. |
| 1.75 | +/- | -NaN |
| -0.74 | +/- | 0. |
| -0.74 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.08 | +/- | 0. |
| 4.55 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -1.70 |
| -9999.00 |

SMC6
| 3613. | +/- | 1. |
| 3750. | +/- | 69. |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| 2.54 | +/- | 0. |
| 2.54 | +/- | -NaN |
| -1.14 | +/- | 0. |
| -1.14 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.97 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4190. | +/- | 9. |
| 4319. | +/- | 112. |
| 1.67 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.96 | +/- | 0. |
| -0.96 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 5.18 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.36 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.63 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
SMC6
| 7872. | +/- | 10. |
| 7773. | +/- | 283. |
| 3.67 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.57 | +/- | 1. |
| 0.57 | +/- | -NaN |
| -2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.00 | +/- | 1. |
| 0.00 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.85 | +/- | 0. |
| 0.45 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.25 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 6558. | +/- | 101. |
| 6431. | +/- | 214. |
| 4.46 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.55 | +/- | 0. |
| 0.55 | +/- | -NaN |
| -0.56 | +/- | 0. |
| -0.55 | +/- | 0. |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| -0.03 | +/- | 3. |
| -0.03 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.33 | +/- | 0. |
| 12.60 | +/- | 0. |
| 1.10 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.56 |
| -9999.00 |
SMC6
| 3768. | +/- | 3. |
| 3909. | +/- | 90. |
| -0.02 | +/- | 0. |
| -0.00 | +/- | 0. |
| 2.08 | +/- | 0. |
| 2.08 | +/- | -NaN |
| -1.24 | +/- | 0. |
| -1.24 | +/- | 0. |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.24 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.09 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.22 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 3735. | +/- | 3. |
| 3866. | +/- | 94. |
| 0.32 | +/- | 0. |
| 0.31 | +/- | 0. |
| 2.78 | +/- | 0. |
| 2.78 | +/- | -NaN |
| -0.99 | +/- | 0. |
| -0.99 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.07 | +/- | 0. |
| 5.29 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -2.00 |
| -9999.00 |
BRIGHT_NEIGHBOR
SMC6
| 3966. | +/- | 4. |
| 4103. | +/- | 100. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | 0. |
| 2.05 | +/- | 0. |
| 2.05 | +/- | -NaN |
| -1.13 | +/- | 0. |
| -1.13 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.72 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.11 |
| -9999.00 |
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 4361. | +/- | 29. |
| 4500. | +/- | 125. |
| 1.72 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.92 | +/- | 0. |
| 1.92 | +/- | -NaN |
| -1.20 | +/- | 0. |
| -1.20 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.23 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.10 | +/- | 0. |
| 5.96 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.35 |
| -9999.00 |

SMC6
| 4162. | +/- | 7. |
| 4293. | +/- | 106. |
| 1.52 | +/- | 0. |
| 1.46 | +/- | 0. |
| 2.23 | +/- | 0. |
| 2.23 | +/- | -NaN |
| -1.00 | +/- | 0. |
| -1.00 | +/- | 0. |
| -0.46 | +/- | 0. |
| -0.46 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.07 | +/- | 0. |
| 5.31 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.11 |
| -9999.00 |
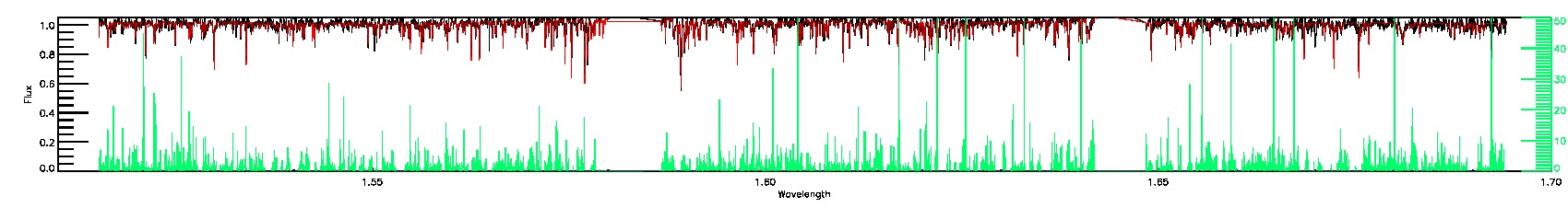
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4168. | +/- | 8. |
| 4301. | +/- | 111. |
| 1.31 | +/- | 0. |
| 1.29 | +/- | 0. |
| 1.95 | +/- | 0. |
| 1.95 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -1.03 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.14 | +/- | 0. |
| 5.39 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.61 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4005. | +/- | 5. |
| 4139. | +/- | 103. |
| 1.16 | +/- | 0. |
| 1.15 | +/- | 0. |
| 2.14 | +/- | 0. |
| 2.14 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -1.08 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.39 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 5.56 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.02 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 4185. | +/- | 7. |
| 4310. | +/- | 107. |
| 1.59 | +/- | 0. |
| 1.50 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -0.85 | +/- | 0. |
| -0.85 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.28 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.87 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.77 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4421. | +/- | 22. |
| 4551. | +/- | 122. |
| 2.26 | +/- | 0. |
| 2.15 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.69 | +/- | -NaN |
| -0.96 | +/- | 0. |
| -0.96 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.29 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 5.17 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.97 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4007. | +/- | 7. |
| 4143. | +/- | 106. |
| 1.18 | +/- | 0. |
| 1.17 | +/- | 0. |
| 2.27 | +/- | 0. |
| 2.27 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -1.11 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.06 | +/- | 0. |
| 5.66 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.10 |
| -9999.00 |

SMC6
| 3695. | +/- | 1. |
| 3830. | +/- | 71. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | 0. |
| 2.82 | +/- | 0. |
| 2.82 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -1.11 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.12 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.66 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -2.00 |
| -9999.00 |
SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 5802. | +/- | 215. |
| -10000. | +/- | -NaN |
| 3.93 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.72 | +/- | 4. |
| 1.72 | +/- | -NaN |
| -2.41 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 3. |
| -9999.99 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 83.29 | +/- | 0. |
| 1.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.90 |
| -9999.00 |

SMC6
| 6493. | +/- | 17. |
| 6357. | +/- | 145. |
| 4.66 | +/- | 0. |
| 4.49 | +/- | 0. |
| 1.04 | +/- | 0. |
| 1.04 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.20 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.86 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.27 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4245. | +/- | 13. |
| 4388. | +/- | 117. |
| 1.59 | +/- | 0. |
| 1.56 | +/- | 0. |
| 2.05 | +/- | 0. |
| 2.05 | +/- | -NaN |
| -1.27 | +/- | 0. |
| -1.27 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 6.23 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.13 |
| -9999.00 |

STAR_BAD
SMC6
| 3603. | +/- | 1. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.48 | +/- | 0. |
| 2.48 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.41 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.64 |
| -9999.00 |
SMC6
| 3989. | +/- | 3. |
| 4117. | +/- | 95. |
| 0.98 | +/- | 0. |
| 0.95 | +/- | 0. |
| 2.01 | +/- | 0. |
| 2.01 | +/- | -NaN |
| -0.94 | +/- | 0. |
| -0.93 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.11 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.90 |
| -9999.00 |

STAR_BAD
SMC6
| 4000. | +/- | 3. |
| -10000. | +/- | -NaN |
| 1.46 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.94 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.81 |
| -9999.00 |
SMC6
| 3901. | +/- | 3. |
| 4033. | +/- | 95. |
| 0.86 | +/- | 0. |
| 0.84 | +/- | 0. |
| 2.13 | +/- | 0. |
| 2.13 | +/- | -NaN |
| -1.01 | +/- | 0. |
| -1.01 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.34 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4095. | +/- | 10. |
| 4229. | +/- | 111. |
| 1.06 | +/- | 0. |
| 1.05 | +/- | 0. |
| 2.45 | +/- | 0. |
| 2.45 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.50 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.47 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| -1.69 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4313. | +/- | 21. |
| 4457. | +/- | 123. |
| 1.20 | +/- | 0. |
| 1.22 | +/- | 0. |
| 2.49 | +/- | 0. |
| 2.49 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -1.29 | +/- | 0. |
| -0.74 | +/- | 0. |
| -0.74 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 6.30 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.79 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| 0.22 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4357. | +/- | 19. |
| 4483. | +/- | 120. |
| 1.71 | +/- | 0. |
| 1.61 | +/- | 0. |
| 2.03 | +/- | 0. |
| 2.03 | +/- | -NaN |
| -0.87 | +/- | 0. |
| -0.87 | +/- | 0. |
| -0.38 | +/- | 0. |
| -0.38 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 4.92 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.90 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4090. | +/- | 8. |
| 4228. | +/- | 108. |
| 1.06 | +/- | 0. |
| 1.06 | +/- | 0. |
| 2.03 | +/- | 0. |
| 2.03 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -1.15 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 5.78 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.94 |
| -9999.00 |
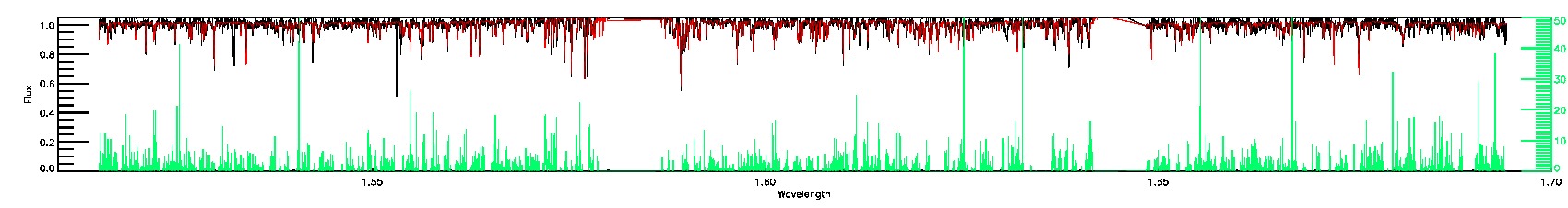
STAR_WARN,SN_WARN
SMC6
| 4432. | +/- | 13. |
| 4561. | +/- | 115. |
| 1.92 | +/- | 0. |
| 1.81 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.83 | +/- | -NaN |
| -0.94 | +/- | 0. |
| -0.94 | +/- | 0. |
| -0.41 | +/- | 0. |
| -0.41 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.13 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.82 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.15 |
| -9999.00 |

STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
SMC6
| 3682. | +/- | 16. |
| -10000. | +/- | -NaN |
| 4.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.11 | +/- | 0. |
| 1.11 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.87 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.97 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4372. | +/- | 23. |
| 4497. | +/- | 121. |
| 2.63 | +/- | 0. |
| 2.52 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -0.86 | +/- | 0. |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.06 | +/- | 0. |
| 4.88 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.88 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.97 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.67 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4043. | +/- | 8. |
| 4175. | +/- | 108. |
| 1.13 | +/- | 0. |
| 1.11 | +/- | 0. |
| 2.11 | +/- | 0. |
| 2.11 | +/- | -NaN |
| -1.03 | +/- | 0. |
| -1.03 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 5.42 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.09 |
| -9999.00 |

STAR_BAD
SMC6
| 3689. | +/- | 2. |
| -10000. | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.56 | +/- | 0. |
| 2.56 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.79 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.91 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4253. | +/- | 9. |
| 4377. | +/- | 112. |
| 1.64 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.83 | +/- | 0. |
| -0.83 | +/- | 0. |
| -0.23 | +/- | 0. |
| -0.23 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.07 | +/- | 0. |
| 4.80 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.12 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 5914. | +/- | 82. |
| 5849. | +/- | 171. |
| 4.44 | +/- | 0. |
| 4.42 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.97 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.20 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |

SMC6
| 6337. | +/- | 20. |
| 6222. | +/- | 139. |
| 4.50 | +/- | 0. |
| 4.39 | +/- | 0. |
| 1.01 | +/- | 0. |
| 1.01 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 8.41 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.05 |
| -9999.00 |
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 4234. | +/- | 12. |
| 4378. | +/- | 116. |
| 1.19 | +/- | 0. |
| 1.22 | +/- | 0. |
| 2.27 | +/- | 0. |
| 2.27 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -1.29 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.04 | +/- | 0. |
| 6.31 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.49 |
| -9999.00 |

STAR_BAD
SMC6
| 3569. | +/- | 1. |
| -10000. | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.94 | +/- | 0. |
| 1.94 | +/- | -NaN |
| -1.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.36 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.69 |
| -9999.00 |
SMC6
| 4003. | +/- | 3. |
| 4124. | +/- | 96. |
| 1.24 | +/- | 0. |
| 1.19 | +/- | 0. |
| 1.96 | +/- | 0. |
| 1.96 | +/- | -NaN |
| -0.76 | +/- | 0. |
| -0.76 | +/- | 0. |
| -0.25 | +/- | 0. |
| -0.25 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.60 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_BAD
SMC6
| 3998. | +/- | 3. |
| -10000. | +/- | -NaN |
| 1.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.91 | +/- | 0. |
| 1.91 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.66 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -2.03 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 4263. | +/- | 13. |
| 4406. | +/- | 119. |
| 1.50 | +/- | 0. |
| 1.48 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.58 | +/- | -NaN |
| -1.27 | +/- | 0. |
| -1.26 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.13 | +/- | 0. |
| 6.20 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.16 |
| -9999.00 |

SMC6
| 4904. | +/- | 18. |
| 4976. | +/- | 124. |
| 3.01 | +/- | 0. |
| 2.63 | +/- | 0. |
| 1.03 | +/- | 0. |
| 1.03 | +/- | -NaN |
| -0.71 | +/- | 0. |
| -0.71 | +/- | 0. |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.23 | +/- | 0. |
| 4.48 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.23 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.58 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4198. | +/- | 14. |
| 4342. | +/- | 117. |
| 1.17 | +/- | 0. |
| 1.20 | +/- | 0. |
| 1.71 | +/- | 0. |
| 1.71 | +/- | -NaN |
| -1.31 | +/- | 0. |
| -1.30 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.47 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 6.35 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.18 |
| -9999.00 |

SMC6
| 4033. | +/- | 6. |
| 4205. | +/- | 111. |
| 0.31 | +/- | 0. |
| 0.42 | +/- | 0. |
| 2.39 | +/- | 0. |
| 2.39 | +/- | -NaN |
| -1.94 | +/- | 0. |
| -1.94 | +/- | 0. |
| -0.74 | +/- | 0. |
| -0.74 | +/- | -NaN |
| -0.38 | +/- | 0. |
| -0.38 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 9.18 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.76 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -2.29 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 4275. | +/- | 33. |
| 4468. | +/- | 139. |
| 0.76 | +/- | 0. |
| 0.93 | +/- | 0. |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| -2.45 | +/- | 0. |
| -2.45 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.28 | +/- | -NaN |
| -0.06 | +/- | 1. |
| -0.06 | +/- | -NaN |
| 0.54 | +/- | 0. |
| 0.50 | +/- | 0. |
| 12.38 | +/- | 0. |
| 1.09 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 1.12 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -2.45 |
| -9999.00 |

SMC6
| 3555. | +/- | 3. |
| 3699. | +/- | 88. |
| 0.16 | +/- | 0. |
| 0.18 | +/- | 0. |
| 2.98 | +/- | 0. |
| 2.98 | +/- | -NaN |
| -1.30 | +/- | 0. |
| -1.30 | +/- | 0. |
| -0.38 | +/- | 0. |
| -0.38 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.14 | +/- | 0. |
| 6.33 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.99 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -2.38 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 4232. | +/- | 16. |
| 4365. | +/- | 117. |
| 2.27 | +/- | 0. |
| 2.17 | +/- | 0. |
| 1.99 | +/- | 0. |
| 1.99 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.44 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.47 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.35 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.04 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4074. | +/- | 7. |
| 4212. | +/- | 106. |
| 1.13 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.66 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -1.18 | +/- | 0. |
| -0.49 | +/- | 0. |
| -0.49 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.88 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.48 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.97 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4164. | +/- | 7. |
| 4295. | +/- | 109. |
| 1.63 | +/- | 0. |
| 1.55 | +/- | 0. |
| 1.58 | +/- | 0. |
| 1.58 | +/- | -NaN |
| -0.99 | +/- | 0. |
| -0.99 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.14 | +/- | 0. |
| 5.29 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.84 |
| -9999.00 |

BRIGHT_NEIGHBOR
SMC6
| 3938. | +/- | 3. |
| 4071. | +/- | 99. |
| 0.92 | +/- | 0. |
| 0.91 | +/- | 0. |
| 2.24 | +/- | 0. |
| 2.24 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -1.06 | +/- | 0. |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.50 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -2.00 |
| -9999.00 |
SMC6
| 6714. | +/- | 42. |
| 6547. | +/- | 194. |
| 4.58 | +/- | 0. |
| 4.31 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.43 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 14.52 | +/- | 0. |
| 1.16 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.77 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.38 |
| -9999.00 |
STAR_BAD,CHI2_BAD,SN_BAD
STAR_WARN,CHI2_WARN,COLORTE_WARN,SN_WARN
SMC6
| 5489. | +/- | 134. |
| -10000. | +/- | -NaN |
| 4.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.01 | +/- | 0. |
| 2.01 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.31 | +/- | 0. |
| 0.86 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| 0.62 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
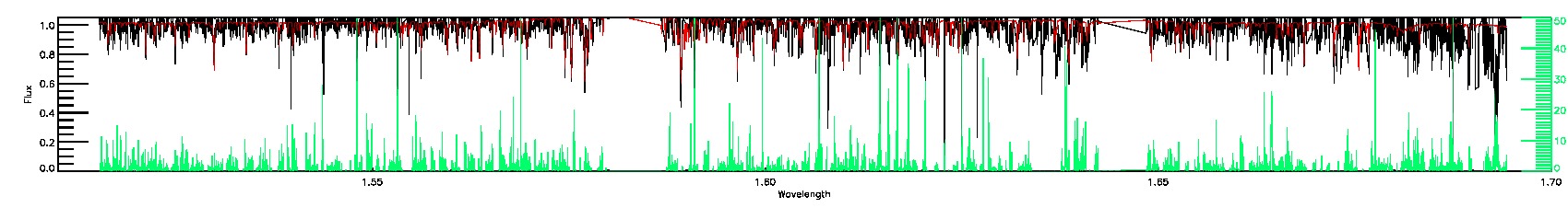
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4147. | +/- | 11. |
| 4288. | +/- | 114. |
| 1.17 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.87 | +/- | 0. |
| 1.87 | +/- | -NaN |
| -1.23 | +/- | 0. |
| -1.23 | +/- | 0. |
| -0.52 | +/- | 0. |
| -0.52 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 6.08 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.67 |
| -9999.00 |

SMC6
| 3897. | +/- | 3. |
| 4035. | +/- | 96. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | 0. |
| 2.09 | +/- | 0. |
| 2.09 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -1.15 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.59 | +/- | 0. |
| 0.59 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.14 | +/- | 0. |
| 5.78 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -2.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.14 |
| -9999.00 |
STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN
SMC6
| 3134. | +/- | 2. |
| -10000. | +/- | -NaN |
| 3.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.48 | +/- | 0. |
| 3.48 | +/- | -NaN |
| 0.42 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.66 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.17 |
| -9999.00 |
SUSPECT_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 4242. | +/- | 21. |
| 4429. | +/- | 133. |
| 0.70 | +/- | 0. |
| 0.86 | +/- | 0. |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| -2.31 | +/- | 0. |
| -2.31 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.09 | +/- | 1. |
| 0.09 | +/- | -NaN |
| 0.54 | +/- | 0. |
| 0.50 | +/- | 0. |
| 11.41 | +/- | 0. |
| 1.06 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.42 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.10 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4197. | +/- | 7. |
| 4324. | +/- | 108. |
| 1.44 | +/- | 0. |
| 1.38 | +/- | 0. |
| 2.09 | +/- | 0. |
| 2.09 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -0.90 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 5.00 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.12 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4168. | +/- | 6. |
| 4290. | +/- | 104. |
| 1.54 | +/- | 0. |
| 1.45 | +/- | 0. |
| 2.17 | +/- | 0. |
| 2.17 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -0.78 | +/- | 0. |
| -0.30 | +/- | 0. |
| -0.30 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 4.67 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| -1.71 |
| -9999.00 |

SMC6
| 5307. | +/- | 17. |
| 5302. | +/- | 123. |
| 4.00 | +/- | 0. |
| 3.99 | +/- | 0. |
| 0.81 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.94 | +/- | 0. |
| 0.47 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.60 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.43 |
| -9999.00 |

SMC6
| 4007. | +/- | 4. |
| 4142. | +/- | 100. |
| 1.12 | +/- | 0. |
| 1.12 | +/- | 0. |
| 2.19 | +/- | 0. |
| 2.19 | +/- | -NaN |
| -1.10 | +/- | 0. |
| -1.10 | +/- | 0. |
| -0.41 | +/- | 0. |
| -0.41 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.16 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.62 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.37 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4190. | +/- | 13. |
| 4316. | +/- | 115. |
| 1.77 | +/- | 0. |
| 1.66 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.88 | +/- | 0. |
| -0.88 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.95 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.98 |
| -9999.00 |
SMC6
| 3890. | +/- | 3. |
| 4027. | +/- | 95. |
| 0.86 | +/- | 0. |
| 0.86 | +/- | 0. |
| 2.31 | +/- | 0. |
| 2.31 | +/- | -NaN |
| -1.13 | +/- | 0. |
| -1.13 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.44 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.72 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.81 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4474. | +/- | 26. |
| 4620. | +/- | 126. |
| 1.46 | +/- | 0. |
| 1.45 | +/- | 0. |
| 2.23 | +/- | 0. |
| 2.23 | +/- | -NaN |
| -1.35 | +/- | 0. |
| -1.35 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.50 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 6.50 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.38 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.45 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4073. | +/- | 12. |
| 4218. | +/- | 114. |
| 1.04 | +/- | 0. |
| 1.07 | +/- | 0. |
| 2.33 | +/- | 0. |
| 2.33 | +/- | -NaN |
| -1.34 | +/- | 0. |
| -1.34 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 6.47 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.95 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -2.31 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -2.27 |
| -9999.00 |

SMC6
| 6353. | +/- | 18. |
| 6232. | +/- | 139. |
| 4.49 | +/- | 0. |
| 4.35 | +/- | 0. |
| 1.51 | +/- | 0. |
| 1.51 | +/- | -NaN |
| -0.15 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.11 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 8.06 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.25 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4351. | +/- | 16. |
| 4472. | +/- | 117. |
| 2.01 | +/- | 0. |
| 1.88 | +/- | 0. |
| 1.33 | +/- | 0. |
| 1.33 | +/- | -NaN |
| -0.76 | +/- | 0. |
| -0.76 | +/- | 0. |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.60 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.63 |
| -9999.00 |

BAD_PIXELS,LOW_SNR,PERSIST_JUMP_POS,SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 4401. | +/- | 20. |
| 4520. | +/- | 120. |
| 3.35 | +/- | 0. |
| 3.28 | +/- | 0. |
| 0.41 | +/- | 0. |
| 0.41 | +/- | -NaN |
| -0.72 | +/- | 0. |
| -0.72 | +/- | 0. |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.25 | +/- | 0. |
| 19.18 | +/- | 0. |
| 1.28 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.08 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.46 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4139. | +/- | 6. |
| 4261. | +/- | 104. |
| 1.47 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.77 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -0.78 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.03 | +/- | 0. |
| 4.68 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.48 |
| -9999.00 |

SMC6
| 4920. | +/- | 10. |
| 4959. | +/- | 108. |
| 4.56 | +/- | 0. |
| 4.62 | +/- | 0. |
| 0.99 | +/- | 0. |
| 0.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.06 | +/- | 0. |
| 1.61 | +/- | 0. |
| 0.21 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.00 |
| -9999.00 |

SMC6
| 5553. | +/- | 20. |
| 5534. | +/- | 129. |
| 4.52 | +/- | 0. |
| 4.61 | +/- | 0. |
| 1.02 | +/- | 0. |
| 1.02 | +/- | -NaN |
| -0.35 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.09 | +/- | 0. |
| 1.67 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -0.54 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 3964. | +/- | 5. |
| 4102. | +/- | 104. |
| 0.85 | +/- | 0. |
| 0.86 | +/- | 0. |
| 2.31 | +/- | 0. |
| 2.31 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -1.16 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.84 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.85 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -2.13 |
| -9999.00 |
| -2.09 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4106. | +/- | 9. |
| 4247. | +/- | 109. |
| 1.40 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.96 | +/- | 0. |
| 1.96 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -1.22 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.47 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.06 | +/- | 0. |
| 6.05 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -2.16 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.17 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4323. | +/- | 19. |
| 4463. | +/- | 123. |
| 1.68 | +/- | 0. |
| 1.63 | +/- | 0. |
| 1.49 | +/- | 0. |
| 1.49 | +/- | -NaN |
| -1.22 | +/- | 0. |
| -1.22 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| 0.28 | +/- | 0. |
| 0.25 | +/- | 0. |
| 6.03 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.34 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.93 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -0.23 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4033. | +/- | 12. |
| 4171. | +/- | 111. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | 0. |
| 2.04 | +/- | 0. |
| 2.04 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -1.15 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.17 | +/- | 0. |
| 5.80 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.91 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4217. | +/- | 7. |
| 4347. | +/- | 109. |
| 1.42 | +/- | 0. |
| 1.37 | +/- | 0. |
| 2.07 | +/- | 0. |
| 2.07 | +/- | -NaN |
| -0.97 | +/- | 0. |
| -0.97 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.22 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.83 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4130. | +/- | 10. |
| 4259. | +/- | 111. |
| 1.60 | +/- | 0. |
| 1.52 | +/- | 0. |
| 1.78 | +/- | 0. |
| 1.78 | +/- | -NaN |
| -0.93 | +/- | 0. |
| -0.93 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 5.11 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.85 |
| -9999.00 |

LOW_SNR
STAR_WARN,SN_WARN
SMC6
| 4926. | +/- | 32. |
| 4965. | +/- | 127. |
| 4.72 | +/- | 0. |
| 4.78 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 2.63 | +/- | 0. |
| 0.42 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.63 |
| -9999.00 |

SMC6
| 3724. | +/- | 2. |
| 3864. | +/- | 90. |
| 0.07 | +/- | 0. |
| 0.08 | +/- | 0. |
| 2.11 | +/- | 0. |
| 2.11 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -1.21 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.27 | +/- | 0. |
| 0.27 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 6.01 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.52 |
| -9999.00 |
SMC6
| 4776. | +/- | 7. |
| 4857. | +/- | 97. |
| 2.57 | +/- | 0. |
| 2.31 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.83 | +/- | -NaN |
| -0.58 | +/- | 0. |
| -0.57 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.21 | +/- | 0. |
| 4.14 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.24 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4285. | +/- | 15. |
| 4418. | +/- | 118. |
| 1.61 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.99 | +/- | 0. |
| 1.99 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.46 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -2.11 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4196. | +/- | 12. |
| 4341. | +/- | 117. |
| 1.15 | +/- | 0. |
| 1.18 | +/- | 0. |
| 2.02 | +/- | 0. |
| 2.02 | +/- | -NaN |
| -1.34 | +/- | 0. |
| -1.34 | +/- | 0. |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 6.46 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.28 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.00 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 3974. | +/- | 9. |
| 4113. | +/- | 106. |
| 0.11 | +/- | 0. |
| 0.11 | +/- | 0. |
| 1.82 | +/- | 0. |
| 1.82 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -1.16 | +/- | 0. |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.08 | +/- | 0. |
| 5.84 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.82 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4083. | +/- | 14. |
| 4224. | +/- | 115. |
| 1.10 | +/- | 0. |
| 1.11 | +/- | 0. |
| 2.37 | +/- | 0. |
| 2.37 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -1.21 | +/- | 0. |
| -0.51 | +/- | 0. |
| -0.51 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.07 | +/- | 0. |
| 6.02 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -2.21 |
| -9999.00 |

SMC6
| 3859. | +/- | 2. |
| 3990. | +/- | 91. |
| 0.74 | +/- | 0. |
| 0.73 | +/- | 0. |
| 2.28 | +/- | 0. |
| 2.28 | +/- | -NaN |
| -0.99 | +/- | 0. |
| -0.99 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.39 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 5.29 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.70 |
| -9999.00 |
SMC6
| 3964. | +/- | 3. |
| 4099. | +/- | 97. |
| 1.09 | +/- | 0. |
| 1.08 | +/- | 0. |
| 2.05 | +/- | 0. |
| 2.05 | +/- | -NaN |
| -1.08 | +/- | 0. |
| -1.08 | +/- | 0. |
| -0.28 | +/- | 0. |
| -0.28 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.04 | +/- | 0. |
| 5.56 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.53 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.92 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4301. | +/- | 18. |
| 4442. | +/- | 121. |
| 1.44 | +/- | 0. |
| 1.43 | +/- | 0. |
| 1.44 | +/- | 0. |
| 1.44 | +/- | -NaN |
| -1.23 | +/- | 0. |
| -1.23 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -0.17 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.08 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -2.34 |
| -9999.00 |
| -2.21 |
| -9999.00 |

SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,ROTATION_WARN,SN_WARN
SMC6
| 4298. | +/- | 34. |
| -10000. | +/- | -NaN |
| 0.49 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 1. |
| 0.98 | +/- | -NaN |
| -2.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 12.76 | +/- | 0. |
| 1.11 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -2.43 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4269. | +/- | 13. |
| 4406. | +/- | 116. |
| 1.60 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.90 | +/- | 0. |
| 1.90 | +/- | -NaN |
| -1.14 | +/- | 0. |
| -1.14 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| 0.18 | +/- | 0. |
| 0.18 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -2.10 |
| -9999.00 |

STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN,COLORTE_WARN
SMC6
| 3336. | +/- | 1. |
| -10000. | +/- | -NaN |
| 4.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.38 | +/- | 0. |
| 4.38 | +/- | -NaN |
| 0.12 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 7.85 | +/- | 0. |
| 0.89 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.91 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.64 |
| -9999.00 |
BRIGHT_NEIGHBOR
SMC6
| 5065. | +/- | 16. |
| 5122. | +/- | 120. |
| 2.73 | +/- | 0. |
| 2.41 | +/- | 0. |
| 1.56 | +/- | 0. |
| 1.56 | +/- | -NaN |
| -0.80 | +/- | 0. |
| -0.80 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.29 | +/- | 0. |
| 0.26 | +/- | 0. |
| 4.71 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.18 |
| -9999.00 |

SMC6
| 4151. | +/- | 6. |
| 4281. | +/- | 104. |
| 1.36 | +/- | 0. |
| 1.32 | +/- | 0. |
| 2.06 | +/- | 0. |
| 2.06 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -0.98 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.26 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.26 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.37 |
| -9999.00 |

BRIGHT_NEIGHBOR
SMC6
| 3943. | +/- | 4. |
| 4077. | +/- | 99. |
| 1.00 | +/- | 0. |
| 0.99 | +/- | 0. |
| 1.91 | +/- | 0. |
| 1.91 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -1.06 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.39 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.03 | +/- | 0. |
| 5.49 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -0.49 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4102. | +/- | 12. |
| 4270. | +/- | 116. |
| 0.55 | +/- | 0. |
| 0.65 | +/- | 0. |
| 2.54 | +/- | 0. |
| 2.54 | +/- | -NaN |
| -1.86 | +/- | 0. |
| -1.86 | +/- | 0. |
| -1.08 | +/- | 0. |
| -1.08 | +/- | -NaN |
| 0.71 | +/- | 0. |
| 0.71 | +/- | -NaN |
| -0.20 | +/- | 0. |
| -0.23 | +/- | 0. |
| 8.80 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.05 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.71 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.09 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_RV_COMBINATION
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 8593. | +/- | 153. |
| 8471. | +/- | 456. |
| 3.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.00 | +/- | 0. |
| 10.00 | +/- | -NaN |
| -1.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| 0.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 10.31 | +/- | 1. |
| 1.01 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -2.26 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.88 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.02 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -1.88 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 6157. | +/- | 140. |
| 6067. | +/- | 192. |
| 5.22 | +/- | 0. |
| 5.18 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.17 | +/- | -NaN |
| -0.32 | +/- | 0. |
| -0.32 | +/- | 0. |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.97 | +/- | 0. |
| 0.97 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 15.23 | +/- | 0. |
| 1.18 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| -1.86 |
| -9999.00 |
| -0.31 |
| -9999.00 |
BRIGHT_NEIGHBOR
SMC6
| 3900. | +/- | 4. |
| 4039. | +/- | 100. |
| 0.75 | +/- | 0. |
| 0.76 | +/- | 0. |
| 2.38 | +/- | 0. |
| 2.38 | +/- | -NaN |
| -1.19 | +/- | 0. |
| -1.19 | +/- | 0. |
| -0.40 | +/- | 0. |
| -0.40 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.05 | +/- | 0. |
| 5.93 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.40 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -1.00 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_WARN,ROTATION_WARN,SN_WARN
SMC6
| 4264. | +/- | 12. |
| 4393. | +/- | 116. |
| 2.15 | +/- | 0. |
| 2.04 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.05 | +/- | -NaN |
| -0.96 | +/- | 0. |
| -0.96 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 5.17 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.05 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.84 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4492. | +/- | 18. |
| 4619. | +/- | 120. |
| 1.82 | +/- | 0. |
| 1.71 | +/- | 0. |
| 2.19 | +/- | 0. |
| 2.19 | +/- | -NaN |
| -0.90 | +/- | 0. |
| -0.90 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.00 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.16 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4397. | +/- | 22. |
| 4525. | +/- | 121. |
| 1.80 | +/- | 0. |
| 1.70 | +/- | 0. |
| 1.78 | +/- | 0. |
| 1.78 | +/- | -NaN |
| -0.93 | +/- | 0. |
| -0.93 | +/- | 0. |
| -0.31 | +/- | 0. |
| -0.31 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.14 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.10 | +/- | 0. |
| 0.71 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.37 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.86 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.25 |
| -9999.00 |

SMC6
| 4084. | +/- | 6. |
| 4221. | +/- | 104. |
| 1.18 | +/- | 0. |
| 1.18 | +/- | 0. |
| 1.92 | +/- | 0. |
| 1.92 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -1.15 | +/- | 0. |
| -0.41 | +/- | 0. |
| -0.41 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.00 | +/- | 0. |
| 5.79 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.30 |
| -9999.00 |

SMC6
| 6291. | +/- | 17. |
| 6167. | +/- | 132. |
| 4.66 | +/- | 0. |
| 4.43 | +/- | 0. |
| 0.84 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.03 | +/- | 0. |
| 6.97 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
SMC6
| 3796. | +/- | 2. |
| 3929. | +/- | 92. |
| 0.49 | +/- | 0. |
| 0.48 | +/- | 0. |
| 2.45 | +/- | 0. |
| 2.45 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -1.06 | +/- | 0. |
| -0.47 | +/- | 0. |
| -0.47 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 5.50 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.46 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.91 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4102. | +/- | 8. |
| 4240. | +/- | 108. |
| 1.27 | +/- | 0. |
| 1.27 | +/- | 0. |
| 2.11 | +/- | 0. |
| 2.11 | +/- | -NaN |
| -1.15 | +/- | 0. |
| -1.15 | +/- | 0. |
| -0.44 | +/- | 0. |
| -0.44 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.02 | +/- | 0. |
| 5.81 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.87 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -2.24 |
| -9999.00 |

SMC6
| 4242. | +/- | 1. |
| 4356. | +/- | 78. |
| 1.99 | +/- | 0. |
| 1.84 | +/- | 0. |
| 1.59 | +/- | 0. |
| 1.59 | +/- | -NaN |
| -0.61 | +/- | 0. |
| -0.61 | +/- | 0. |
| 0.04 | +/- | 0. |
| 0.04 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.23 | +/- | 0. |
| 0.19 | +/- | 0. |
| 4.22 | +/- | 0. |
| 0.63 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.72 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -0.51 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.58 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_WARN,ROTATION_WARN,SN_WARN
SMC6
| 4201. | +/- | 12. |
| 4362. | +/- | 121. |
| 1.02 | +/- | 0. |
| 1.09 | +/- | 0. |
| 0.39 | +/- | 0. |
| 0.39 | +/- | -NaN |
| -1.72 | +/- | 0. |
| -1.72 | +/- | 0. |
| -0.48 | +/- | 0. |
| -0.48 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.27 | +/- | 0. |
| 8.07 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.75 |
| -9999.00 |
| -2.46 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.15 |
| -9999.00 |

BRIGHT_NEIGHBOR,VERY_BRIGHT_NEIGHBOR
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 4004. | +/- | 7. |
| 4142. | +/- | 106. |
| 0.99 | +/- | 0. |
| 1.00 | +/- | 0. |
| 1.99 | +/- | 0. |
| 1.99 | +/- | -NaN |
| -1.18 | +/- | 0. |
| -1.18 | +/- | 0. |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 5.91 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.41 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.15 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4485. | +/- | 10. |
| 4575. | +/- | 106. |
| 4.53 | +/- | 0. |
| 4.69 | +/- | 0. |
| 0.76 | +/- | 0. |
| 0.76 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 3.50 | +/- | 0. |
| 0.54 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.74 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.24 |
| -9999.00 |

SMC6
| 4100. | +/- | 5. |
| 4224. | +/- | 102. |
| 1.38 | +/- | 0. |
| 1.32 | +/- | 0. |
| 2.01 | +/- | 0. |
| 2.01 | +/- | -NaN |
| -0.84 | +/- | 0. |
| -0.84 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.03 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.83 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.84 |
| -9999.00 |

SMC6
| 6342. | +/- | 15. |
| 6209. | +/- | 133. |
| 4.58 | +/- | 0. |
| 4.31 | +/- | 0. |
| 0.94 | +/- | 0. |
| 0.94 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.17 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| -0.21 | +/- | 0. |
| -0.21 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 9.63 | +/- | 0. |
| 0.98 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.64 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.15 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4219. | +/- | 11. |
| 4359. | +/- | 117. |
| 1.26 | +/- | 0. |
| 1.27 | +/- | 0. |
| 2.34 | +/- | 0. |
| 2.34 | +/- | -NaN |
| -1.21 | +/- | 0. |
| -1.21 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.15 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 6.00 | +/- | 0. |
| 0.78 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -2.19 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| -2.18 |
| -9999.00 |
| -0.65 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4168. | +/- | 6. |
| 4290. | +/- | 105. |
| 1.46 | +/- | 0. |
| 1.38 | +/- | 0. |
| 1.90 | +/- | 0. |
| 1.90 | +/- | -NaN |
| -0.77 | +/- | 0. |
| -0.77 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.30 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.25 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4157. | +/- | 16. |
| 4300. | +/- | 118. |
| 1.34 | +/- | 0. |
| 1.35 | +/- | 0. |
| 2.14 | +/- | 0. |
| 2.14 | +/- | -NaN |
| -1.28 | +/- | 0. |
| -1.28 | +/- | 0. |
| -0.70 | +/- | 0. |
| -0.70 | +/- | -NaN |
| 0.77 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.25 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 1.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -2.21 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -2.50 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4246. | +/- | 12. |
| 4367. | +/- | 113. |
| 1.82 | +/- | 0. |
| 1.69 | +/- | 0. |
| 1.90 | +/- | 0. |
| 1.90 | +/- | -NaN |
| -0.75 | +/- | 0. |
| -0.75 | +/- | 0. |
| -0.36 | +/- | 0. |
| -0.36 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.58 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.71 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.89 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4506. | +/- | 21. |
| 4651. | +/- | 126. |
| 1.65 | +/- | 0. |
| 1.62 | +/- | 0. |
| 1.47 | +/- | 0. |
| 1.47 | +/- | -NaN |
| -1.35 | +/- | 0. |
| -1.35 | +/- | 0. |
| -0.38 | +/- | 0. |
| -0.38 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.14 | +/- | 0. |
| 6.51 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.46 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.29 |
| -9999.00 |

LOW_SNR,PERSIST_JUMP_POS
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
SMC6
| 4821. | +/- | 51. |
| -10000. | +/- | -NaN |
| 4.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.11 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.75 | +/- | 0. |
| 0.57 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.12 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.75 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.35 |
| -9999.00 |

SMC6
| 4174. | +/- | 6. |
| 4312. | +/- | 105. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | 0. |
| 2.22 | +/- | 0. |
| 2.22 | +/- | -NaN |
| -1.16 | +/- | 0. |
| -1.16 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 5.84 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -2.27 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4162. | +/- | 6. |
| 4280. | +/- | 105. |
| 1.70 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.80 | +/- | 0. |
| 1.80 | +/- | -NaN |
| -0.70 | +/- | 0. |
| -0.70 | +/- | 0. |
| -0.26 | +/- | 0. |
| -0.26 | +/- | -NaN |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.01 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.25 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.69 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.68 |
| -9999.00 |

BRIGHT_NEIGHBOR
SMC6
| 5443. | +/- | 20. |
| 5418. | +/- | 132. |
| 4.47 | +/- | 0. |
| 4.40 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | 0. |
| -0.05 | +/- | 0. |
| -0.05 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.05 | +/- | 0. |
| 3.56 | +/- | 0. |
| 0.55 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.25 |
| -9999.00 |

STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN
SMC6
| 3000. | +/- | 0. |
| -10000. | +/- | -NaN |
| 1.47 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.95 | +/- | 0. |
| 2.95 | +/- | -NaN |
| -2.27 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 1.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 11.15 | +/- | 0. |
| 1.05 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.15 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.22 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.26 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.05 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -2.49 |
| -9999.00 |
SUSPECT_RV_COMBINATION,SUSPECT_BROAD_LINES,BAD_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 7615. | +/- | 54. |
| 7440. | +/- | 292. |
| 4.27 | +/- | 0. |
| 4.11 | +/- | 0. |
| 0.92 | +/- | 1. |
| 0.92 | +/- | -NaN |
| -0.62 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.10 | +/- | 1. |
| 0.10 | +/- | -NaN |
| 0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 86.96 | +/- | 0. |
| 1.94 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| 0.94 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.57 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.98 |
| -9999.00 |
SMC6
| 6573. | +/- | 18. |
| 6423. | +/- | 147. |
| 4.55 | +/- | 0. |
| 4.33 | +/- | 0. |
| 1.08 | +/- | 0. |
| 1.08 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.14 | +/- | 0. |
| -0.14 | +/- | -NaN |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 9.92 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| 0.03 |
| -9999.00 |
SMC6
| 6473. | +/- | 21. |
| 6334. | +/- | 156. |
| 4.66 | +/- | 0. |
| 4.45 | +/- | 0. |
| 0.86 | +/- | 0. |
| 0.86 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.00 | +/- | 0. |
| 6.41 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.60 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4137. | +/- | 8. |
| 4263. | +/- | 107. |
| 1.37 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.82 | +/- | 0. |
| 1.82 | +/- | -NaN |
| -0.87 | +/- | 0. |
| -0.87 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.06 | +/- | 0. |
| 4.93 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.97 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.56 |
| -9999.00 |

SUSPECT_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 4024. | +/- | 8. |
| 4160. | +/- | 109. |
| 1.16 | +/- | 0. |
| 1.16 | +/- | 0. |
| 1.76 | +/- | 0. |
| 1.76 | +/- | -NaN |
| -1.13 | +/- | 0. |
| -1.13 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.50 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 5.73 | +/- | 0. |
| 0.76 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.64 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4254. | +/- | 11. |
| 4385. | +/- | 114. |
| 1.40 | +/- | 0. |
| 1.36 | +/- | 0. |
| 1.74 | +/- | 0. |
| 1.74 | +/- | -NaN |
| -1.00 | +/- | 0. |
| -1.00 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.08 | +/- | -NaN |
| 0.14 | +/- | 0. |
| 0.10 | +/- | 0. |
| 5.30 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.58 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.83 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -1.08 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4116. | +/- | 9. |
| 4267. | +/- | 111. |
| 1.00 | +/- | 0. |
| 1.05 | +/- | 0. |
| 1.89 | +/- | 0. |
| 1.89 | +/- | -NaN |
| -1.46 | +/- | 0. |
| -1.46 | +/- | 0. |
| -0.49 | +/- | 0. |
| -0.49 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 6.95 | +/- | 0. |
| 0.84 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -1.05 |
| -9999.00 |

LOW_SNR
STAR_BAD,COLORTE_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 5398. | +/- | 90. |
| -10000. | +/- | -NaN |
| 3.89 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.51 | +/- | 0. |
| 0.51 | +/- | -NaN |
| -0.73 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.44 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.10 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.54 | +/- | 0. |
| 0.66 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.25 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| -2.44 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -2.35 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.84 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4120. | +/- | 8. |
| 4254. | +/- | 110. |
| 1.58 | +/- | 0. |
| 1.52 | +/- | 0. |
| 1.93 | +/- | 0. |
| 1.93 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.02 | +/- | -NaN |
| 0.15 | +/- | 0. |
| 0.12 | +/- | 0. |
| 5.46 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.18 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -0.62 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.50 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -1.80 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.98 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4360. | +/- | 9. |
| 4482. | +/- | 110. |
| 1.80 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.89 | +/- | 0. |
| 1.89 | +/- | -NaN |
| -0.77 | +/- | 0. |
| -0.77 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.13 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.64 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.27 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.87 |
| -9999.00 |

VERY_BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4534. | +/- | 21. |
| 4694. | +/- | 130. |
| 1.23 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.42 | +/- | 0. |
| 1.42 | +/- | -NaN |
| -1.78 | +/- | 0. |
| -1.78 | +/- | 0. |
| -0.50 | +/- | 0. |
| -0.50 | +/- | -NaN |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.13 | +/- | 0. |
| 8.38 | +/- | 0. |
| 0.92 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -1.92 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -1.47 |
| -9999.00 |

SMC6
| 3758. | +/- | 2. |
| 3886. | +/- | 87. |
| 0.52 | +/- | 0. |
| 0.50 | +/- | 0. |
| 2.45 | +/- | 0. |
| 2.45 | +/- | -NaN |
| -0.92 | +/- | 0. |
| -0.92 | +/- | 0. |
| -0.39 | +/- | 0. |
| -0.39 | +/- | -NaN |
| 0.06 | +/- | 0. |
| 0.06 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.05 | +/- | 0. |
| 5.06 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.87 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.22 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4121. | +/- | 10. |
| 4265. | +/- | 114. |
| 0.93 | +/- | 0. |
| 0.95 | +/- | 0. |
| 2.83 | +/- | 0. |
| 2.83 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -1.29 | +/- | 0. |
| -0.54 | +/- | 0. |
| -0.54 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 6.29 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.76 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -1.69 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.69 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.45 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4002. | +/- | 5. |
| 4138. | +/- | 103. |
| 1.07 | +/- | 0. |
| 1.07 | +/- | 0. |
| 2.12 | +/- | 0. |
| 2.12 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -1.11 | +/- | 0. |
| -0.48 | +/- | 0. |
| -0.48 | +/- | -NaN |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.07 | +/- | 0. |
| 5.68 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.47 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -1.49 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.17 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.49 |
| -9999.00 |
| -1.51 |
| -9999.00 |

LOW_SNR,PERSIST_JUMP_POS
STAR_WARN,SN_WARN
SMC6
| 5492. | +/- | 80. |
| 5477. | +/- | 156. |
| 4.22 | +/- | 0. |
| 4.29 | +/- | 0. |
| 0.45 | +/- | 0. |
| 0.45 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.27 | +/- | 0. |
| 0.05 | +/- | 0. |
| 0.05 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.17 | +/- | 0. |
| 0.16 | +/- | 0. |
| 4.44 | +/- | 0. |
| 0.65 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.19 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.20 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.50 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4192. | +/- | 14. |
| 4372. | +/- | 127. |
| 0.59 | +/- | 0. |
| 0.73 | +/- | 0. |
| 2.44 | +/- | 0. |
| 2.44 | +/- | -NaN |
| -2.14 | +/- | 0. |
| -2.14 | +/- | 0. |
| -0.43 | +/- | 0. |
| -0.43 | +/- | -NaN |
| 0.31 | +/- | 0. |
| 0.31 | +/- | -NaN |
| 0.38 | +/- | 0. |
| 0.35 | +/- | 0. |
| 10.36 | +/- | 0. |
| 1.02 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.87 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -2.30 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.11 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -2.14 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.30 |
| -9999.00 |

BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_WARN,ROTATION_WARN,SN_WARN
SMC6
| 4234. | +/- | 12. |
| 4358. | +/- | 114. |
| 2.22 | +/- | 0. |
| 2.09 | +/- | 0. |
| 0.72 | +/- | 0. |
| 0.72 | +/- | -NaN |
| -0.82 | +/- | 0. |
| -0.82 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.00 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.04 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.60 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4118. | +/- | 12. |
| 4254. | +/- | 113. |
| 1.33 | +/- | 0. |
| 1.31 | +/- | 0. |
| 1.92 | +/- | 0. |
| 1.92 | +/- | -NaN |
| -1.11 | +/- | 0. |
| -1.11 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -0.02 | +/- | 0. |
| 5.66 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.43 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.48 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -2.09 |
| -9999.00 |
| -2.29 |
| -9999.00 |
| -2.04 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4289. | +/- | 16. |
| 4441. | +/- | 123. |
| 1.13 | +/- | 0. |
| 1.18 | +/- | 0. |
| 2.43 | +/- | 0. |
| 2.43 | +/- | -NaN |
| -1.49 | +/- | 0. |
| -1.49 | +/- | 0. |
| -0.19 | +/- | 0. |
| -0.19 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.21 | +/- | 0. |
| 7.08 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.22 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -1.67 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| -2.20 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -2.42 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4230. | +/- | 7. |
| 4355. | +/- | 108. |
| 1.46 | +/- | 0. |
| 1.39 | +/- | 0. |
| 1.77 | +/- | 0. |
| 1.77 | +/- | -NaN |
| -0.84 | +/- | 0. |
| -0.84 | +/- | 0. |
| -0.42 | +/- | 0. |
| -0.42 | +/- | -NaN |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| 0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 4.83 | +/- | 0. |
| 0.68 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.42 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.41 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.57 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -1.74 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -1.79 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4230. | +/- | 12. |
| 4357. | +/- | 114. |
| 1.78 | +/- | 0. |
| 1.68 | +/- | 0. |
| 1.57 | +/- | 0. |
| 1.57 | +/- | -NaN |
| -0.89 | +/- | 0. |
| -0.89 | +/- | 0. |
| -0.22 | +/- | 0. |
| -0.22 | +/- | -NaN |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.02 | +/- | 0. |
| 4.98 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.24 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -0.88 |
| -9999.00 |
BRIGHT_NEIGHBOR
SMC6
| 3968. | +/- | 3. |
| 4090. | +/- | 94. |
| 1.05 | +/- | 0. |
| 1.00 | +/- | 0. |
| 2.04 | +/- | 0. |
| 2.04 | +/- | -NaN |
| -0.78 | +/- | 0. |
| -0.77 | +/- | 0. |
| -0.33 | +/- | 0. |
| -0.33 | +/- | -NaN |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.04 | +/- | 0. |
| 4.65 | +/- | 0. |
| 0.67 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.33 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.95 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4201. | +/- | 9. |
| 4329. | +/- | 112. |
| 1.30 | +/- | 0. |
| 1.27 | +/- | 0. |
| 2.03 | +/- | 0. |
| 2.03 | +/- | -NaN |
| -0.92 | +/- | 0. |
| -0.92 | +/- | 0. |
| -0.29 | +/- | 0. |
| -0.29 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.10 | +/- | -NaN |
| 0.12 | +/- | 0. |
| 0.09 | +/- | 0. |
| 5.06 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.29 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.86 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.94 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -1.48 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4106. | +/- | 11. |
| 4254. | +/- | 116. |
| 1.00 | +/- | 0. |
| 1.03 | +/- | 0. |
| 2.37 | +/- | 0. |
| 2.37 | +/- | -NaN |
| -1.39 | +/- | 0. |
| -1.39 | +/- | 0. |
| -0.58 | +/- | 0. |
| -0.58 | +/- | -NaN |
| -0.04 | +/- | 0. |
| -0.04 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.13 | +/- | 0. |
| 6.67 | +/- | 0. |
| 0.82 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.81 |
| -9999.00 |
| -1.77 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -1.82 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.78 |
| -9999.00 |
| -1.37 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| -2.24 |
| -9999.00 |

SMC6
| 6111. | +/- | 13. |
| 6004. | +/- | 124. |
| 4.57 | +/- | 0. |
| 4.32 | +/- | 0. |
| 0.96 | +/- | 0. |
| 0.96 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.22 | +/- | 0. |
| -0.09 | +/- | 0. |
| -0.09 | +/- | -NaN |
| 0.21 | +/- | 0. |
| 0.21 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 3.17 | +/- | 0. |
| 0.50 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| 0.47 |
| -9999.00 |
| 0.34 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| 0.18 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4230. | +/- | 10. |
| 4360. | +/- | 112. |
| 1.45 | +/- | 0. |
| 1.40 | +/- | 0. |
| 1.54 | +/- | 0. |
| 1.54 | +/- | -NaN |
| -0.99 | +/- | 0. |
| -0.99 | +/- | 0. |
| -0.35 | +/- | 0. |
| -0.35 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.11 | +/- | 0. |
| 0.07 | +/- | 0. |
| 5.27 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.39 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| -2.27 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.37 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| 0.07 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 4404. | +/- | 51. |
| -10000. | +/- | -NaN |
| 1.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.52 | +/- | 0. |
| 2.52 | +/- | -NaN |
| -1.82 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.51 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.17 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.57 | +/- | 0. |
| 0.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.21 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.31 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -0.77 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| -1.62 |
| -9999.00 |
| 0.54 |
| -9999.00 |
| -0.70 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -1.98 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -2.23 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -2.42 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.31 |
| -9999.00 |

STAR_BAD,CHI2_BAD
STAR_WARN,CHI2_WARN
SMC6
| 3063. | +/- | 1. |
| -10000. | +/- | -NaN |
| 1.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 3.23 | +/- | 0. |
| 3.23 | +/- | -NaN |
| -0.98 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.24 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.15 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.32 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -1.83 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.41 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -1.55 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.21 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.95 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.64 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -1.16 |
| -9999.00 |
SMC6
| 6356. | +/- | 17. |
| 6229. | +/- | 136. |
| 4.40 | +/- | 0. |
| 4.19 | +/- | 0. |
| 0.72 | +/- | 0. |
| 0.72 | +/- | -NaN |
| -0.00 | +/- | 0. |
| 0.00 | +/- | 0. |
| -0.00 | +/- | 0. |
| -0.00 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 15.32 | +/- | 0. |
| 1.19 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.01 |
| -9999.00 |
STAR_WARN,CHI2_WARN
SMC6
| 3056. | +/- | 1. |
| 3198. | +/- | 68. |
| 0.13 | +/- | 0. |
| 0.15 | +/- | 0. |
| 3.40 | +/- | 0. |
| 3.40 | +/- | -NaN |
| -1.25 | +/- | 0. |
| -1.25 | +/- | 0. |
| 0.19 | +/- | 0. |
| 0.19 | +/- | -NaN |
| -0.50 | +/- | 0. |
| -0.50 | +/- | -NaN |
| -0.27 | +/- | 0. |
| -0.30 | +/- | 0. |
| 6.13 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.50 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.33 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.87 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 4001. | +/- | 7. |
| 4143. | +/- | 107. |
| 0.83 | +/- | 0. |
| 0.85 | +/- | 0. |
| 2.28 | +/- | 0. |
| 2.28 | +/- | -NaN |
| -1.27 | +/- | 0. |
| -1.27 | +/- | 0. |
| -0.67 | +/- | 0. |
| -0.67 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| -0.02 | +/- | 0. |
| -0.06 | +/- | 0. |
| 6.21 | +/- | 0. |
| 0.79 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.65 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.96 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -2.25 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| 0.78 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.68 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.50 |
| -9999.00 |

SUSPECT_BROAD_LINES
STAR_BAD
STAR_WARN,COLORTE_WARN
SMC6
| 7770. | +/- | 12. |
| -10000. | +/- | -NaN |
| 4.39 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.79 | +/- | 0. |
| 4.79 | +/- | -NaN |
| -0.23 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.18 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 30.10 | +/- | 0. |
| 1.48 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.56 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.61 |
| -9999.00 |
| 0.19 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.53 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.39 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.54 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -1.22 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 5388. | +/- | 89. |
| 5417. | +/- | 164. |
| 3.82 | +/- | 0. |
| 4.07 | +/- | 0. |
| 0.46 | +/- | 0. |
| 0.46 | +/- | -NaN |
| -1.02 | +/- | 0. |
| -1.02 | +/- | 0. |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| -0.48 | +/- | 0. |
| -0.48 | +/- | -NaN |
| 0.24 | +/- | 0. |
| 0.20 | +/- | 0. |
| 5.38 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -1.07 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.47 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| 0.55 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| -1.04 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| -1.14 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.86 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 5792. | +/- | 97. |
| 5756. | +/- | 172. |
| 4.09 | +/- | 0. |
| 4.23 | +/- | 0. |
| 0.43 | +/- | 0. |
| 0.43 | +/- | -NaN |
| -0.59 | +/- | 0. |
| -0.59 | +/- | 0. |
| 0.07 | +/- | 0. |
| 0.07 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.13 | +/- | 0. |
| 0.12 | +/- | 0. |
| 4.19 | +/- | 0. |
| 0.62 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.41 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| -0.49 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| 0.76 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.48 |
| -9999.00 |

BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4186. | +/- | 14. |
| 4319. | +/- | 115. |
| 1.46 | +/- | 0. |
| 1.41 | +/- | 0. |
| 2.00 | +/- | 0. |
| 2.00 | +/- | -NaN |
| -1.06 | +/- | 0. |
| -1.06 | +/- | 0. |
| -0.52 | +/- | 0. |
| -0.52 | +/- | -NaN |
| 0.49 | +/- | 0. |
| 0.49 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.11 | +/- | 0. |
| 5.48 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.55 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.93 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -0.74 |
| -9999.00 |
| -1.23 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| -0.71 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -2.08 |
| -9999.00 |
| -2.12 |
| -9999.00 |
| -2.02 |
| -9999.00 |

STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 3972. | +/- | 9. |
| 4098. | +/- | 107. |
| 1.21 | +/- | 0. |
| 1.17 | +/- | 0. |
| 1.85 | +/- | 0. |
| 1.85 | +/- | -NaN |
| -0.87 | +/- | 0. |
| -0.87 | +/- | 0. |
| -0.13 | +/- | 0. |
| -0.13 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -0.41 | +/- | -NaN |
| 0.08 | +/- | 0. |
| 0.05 | +/- | 0. |
| 4.92 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.13 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| -1.24 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.65 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.09 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.55 |
| -9999.00 |
LOW_SNR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
SMC6
| 4202. | +/- | 19. |
| -10000. | +/- | -NaN |
| 1.28 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.51 | +/- | 0. |
| 2.51 | +/- | -NaN |
| -1.29 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.55 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.05 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 6.28 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.51 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| 0.58 |
| -9999.00 |
| -1.36 |
| -9999.00 |
| 0.30 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.25 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.42 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -2.45 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 4168. | +/- | 11. |
| 4263. | +/- | 100. |
| 4.52 | +/- | 0. |
| 4.79 | +/- | 0. |
| 0.34 | +/- | 0. |
| 0.34 | +/- | -NaN |
| -0.16 | +/- | 0. |
| -0.16 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.06 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.04 | +/- | 0. |
| 5.87 | +/- | 0. |
| 0.77 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.00 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -0.16 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -0.43 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -0.36 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| 0.23 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.08 |
| -9999.00 |

LOW_SNR
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
SMC6
| 4691. | +/- | 80. |
| -10000. | +/- | -NaN |
| 2.26 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.75 | +/- | 0. |
| 0.75 | +/- | -NaN |
| -1.74 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.61 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| -0.14 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.19 | +/- | 0. |
| 0.91 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.73 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.19 |
| -9999.00 |
| -2.06 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -1.91 |
| -9999.00 |
| -1.89 |
| -9999.00 |
| -2.33 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| -1.90 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| -1.11 |
| -9999.00 |
| 0.11 |
| -9999.00 |
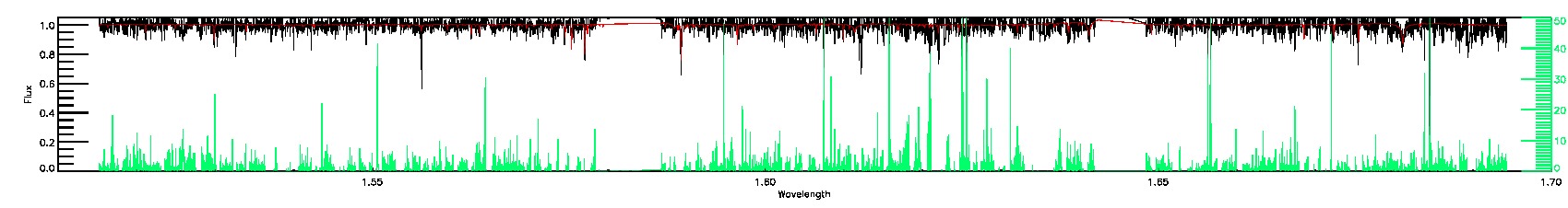
STAR_WARN,SN_WARN
SMC6
| 3809. | +/- | 11. |
| 3911. | +/- | 95. |
| 4.59 | +/- | 0. |
| 5.00 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| -0.31 | +/- | 0. |
| -0.31 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.11 | +/- | 0. |
| -0.11 | +/- | -NaN |
| 0.16 | +/- | 0. |
| 0.15 | +/- | 0. |
| 3.83 | +/- | 0. |
| 0.58 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| 0.32 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.76 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.96 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.63 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.35 |
| -9999.00 |
STAR_BAD,SN_BAD
STAR_WARN,SN_WARN
SMC6
| 4252. | +/- | 18. |
| -10000. | +/- | -NaN |
| 1.34 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 2.47 | +/- | 0. |
| 2.47 | +/- | -NaN |
| -1.09 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.07 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.06 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 5.61 | +/- | 0. |
| 0.75 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.16 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.28 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| -0.58 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.84 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.65 |
| -9999.00 |
| -1.34 |
| -9999.00 |
| -1.07 |
| -9999.00 |
| -1.26 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -1.01 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -0.94 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.04 |
| -9999.00 |

SUSPECT_RV_COMBINATION,BAD_RV_COMBINATION
STAR_WARN,SN_WARN
SMC6
| 6808. | +/- | 61. |
| 6656. | +/- | 228. |
| 4.00 | +/- | 0. |
| 3.97 | +/- | 0. |
| 0.40 | +/- | 0. |
| 0.40 | +/- | -NaN |
| -0.64 | +/- | 0. |
| -0.64 | +/- | 0. |
| 0.01 | +/- | 0. |
| 0.01 | +/- | -NaN |
| 0.01 | +/- | 1. |
| 0.01 | +/- | -NaN |
| 0.78 | +/- | 0. |
| 0.75 | +/- | 0. |
| 84.61 | +/- | 0. |
| 1.93 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.13 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.33 |
| -9999.00 |
| 0.67 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.95 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.80 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -0.73 |
| -9999.00 |
| 0.70 |
| -9999.00 |
| 0.97 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.56 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| -0.14 |
| -9999.00 |
| 0.11 |
| -9999.00 |
| 0.99 |
| -9999.00 |
| 0.66 |
| -9999.00 |
| -0.13 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -2.37 |
| -9999.00 |
BRIGHT_NEIGHBOR,SUSPECT_BROAD_LINES
STAR_WARN,ROTATION_WARN,SN_WARN
SMC6
| 4666. | +/- | 51. |
| 4824. | +/- | 147. |
| 1.87 | +/- | 0. |
| 1.92 | +/- | 0. |
| 0.42 | +/- | 0. |
| 0.42 | +/- | -NaN |
| -2.09 | +/- | 0. |
| -2.09 | +/- | 0. |
| -0.42 | +/- | 1. |
| -0.42 | +/- | -NaN |
| 1.27 | +/- | 1. |
| 1.27 | +/- | -NaN |
| 0.35 | +/- | 0. |
| 0.32 | +/- | 0. |
| 10.07 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 1.43 |
| -9999.00 |
| 0.27 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -2.47 |
| -9999.00 |
| 0.26 |
| -9999.00 |
| -2.48 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.35 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| -1.12 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.37 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -2.24 |
| -9999.00 |
| -2.04 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -2.07 |
| -9999.00 |
| -1.78 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -1.05 |
| -9999.00 |
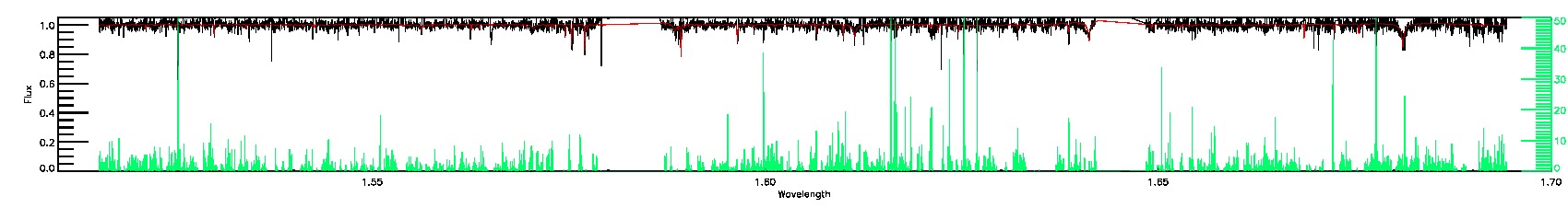
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4020. | +/- | 8. |
| 4149. | +/- | 108. |
| 1.36 | +/- | 0. |
| 1.32 | +/- | 0. |
| 1.85 | +/- | 0. |
| 1.85 | +/- | -NaN |
| -0.97 | +/- | 0. |
| -0.97 | +/- | 0. |
| -0.34 | +/- | 0. |
| -0.34 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -0.03 | +/- | -NaN |
| 0.05 | +/- | 0. |
| 0.01 | +/- | 0. |
| 5.20 | +/- | 0. |
| 0.72 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.05 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -1.52 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.52 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| -2.32 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -1.54 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.81 |
| -9999.00 |
| -0.38 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.44 |
| -9999.00 |
| -2.01 |
| -9999.00 |
| -1.87 |
| -9999.00 |
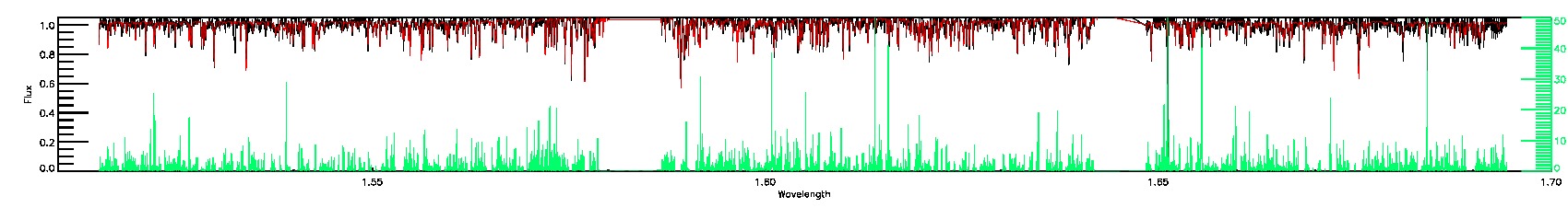
BRIGHT_NEIGHBOR
STAR_WARN,SN_WARN
SMC6
| 4083. | +/- | 6. |
| 4216. | +/- | 106. |
| 1.15 | +/- | 0. |
| 1.14 | +/- | 0. |
| 1.83 | +/- | 0. |
| 1.83 | +/- | -NaN |
| -1.05 | +/- | 0. |
| -1.05 | +/- | 0. |
| -0.32 | +/- | 0. |
| -0.32 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.01 | +/- | -NaN |
| 0.10 | +/- | 0. |
| 0.06 | +/- | 0. |
| 5.46 | +/- | 0. |
| 0.74 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.32 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| -0.68 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| -1.19 |
| -9999.00 |
| 0.17 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.29 |
| -9999.00 |
| -1.00 |
| -9999.00 |
| -1.10 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| -1.02 |
| -9999.00 |
| -1.31 |
| -9999.00 |
| -1.13 |
| -9999.00 |
| -0.61 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -1.03 |
| -9999.00 |
| -0.44 |
| -9999.00 |
| -2.19 |
| -9999.00 |

LOW_SNR
STAR_WARN,SN_WARN
SMC6
| 4255. | +/- | 27. |
| 4433. | +/- | 135. |
| 0.96 | +/- | 0. |
| 1.09 | +/- | 0. |
| 0.32 | +/- | 0. |
| 0.32 | +/- | -NaN |
| -2.08 | +/- | 0. |
| -2.08 | +/- | 0. |
| -0.27 | +/- | 0. |
| -0.27 | +/- | -NaN |
| 0.08 | +/- | 1. |
| 0.08 | +/- | -NaN |
| 0.36 | +/- | 0. |
| 0.32 | +/- | 0. |
| 10.00 | +/- | 0. |
| 1.00 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.45 |
| -9999.00 |
| 0.59 |
| -9999.00 |
| 0.08 |
| -9999.00 |
| 0.44 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.36 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| 0.40 |
| -9999.00 |
| -2.39 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.91 |
| -9999.00 |
| 1.00 |
| -9999.00 |
| 0.28 |
| -9999.00 |
| 0.39 |
| -9999.00 |
| -1.57 |
| -9999.00 |
| -2.38 |
| -9999.00 |
| -1.84 |
| -9999.00 |
| -2.00 |
| -9999.00 |
| -1.38 |
| -9999.00 |
| -1.61 |
| -9999.00 |
| -2.40 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -2.43 |
| -9999.00 |
| -2.36 |
| -9999.00 |
| -0.72 |
| -9999.00 |
| -2.32 |
| -9999.00 |

SMC6
| 3781. | +/- | 2. |
| 3907. | +/- | 88. |
| 0.49 | +/- | 0. |
| 0.45 | +/- | 0. |
| 2.44 | +/- | 0. |
| 2.44 | +/- | -NaN |
| -0.89 | +/- | 0. |
| -0.89 | +/- | 0. |
| -0.24 | +/- | 0. |
| -0.24 | +/- | -NaN |
| 0.09 | +/- | 0. |
| 0.09 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.00 | +/- | 0. |
| 4.97 | +/- | 0. |
| 0.70 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.23 |
| -9999.00 |
| -0.35 |
| -9999.00 |
| 0.10 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.43 |
| -9999.00 |
| 0.07 |
| -9999.00 |
| -1.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.90 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.08 |
| -9999.00 |
| 0.38 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -0.84 |
| -9999.00 |
| -1.18 |
| -9999.00 |
| -0.87 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -0.92 |
| -9999.00 |
| -0.34 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| -0.82 |
| -9999.00 |
| -0.98 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -1.81 |
| -9999.00 |
BRIGHT_NEIGHBOR
STAR_BAD,SN_BAD
STAR_WARN,CHI2_WARN,SN_WARN
SMC6
| 5352. | +/- | 141. |
| -10000. | +/- | -NaN |
| 3.64 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 | +/- | 0. |
| 0.38 | +/- | -NaN |
| -0.86 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.56 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.38 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.40 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 4.89 | +/- | 0. |
| 0.69 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.54 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.50 |
| -9999.00 |
| 0.48 |
| -9999.00 |
| -0.24 |
| -9999.00 |
| 0.45 |
| -9999.00 |
| -0.79 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -2.03 |
| -9999.00 |
| 0.52 |
| -9999.00 |
| 0.89 |
| -9999.00 |
| 0.98 |
| -9999.00 |
| 0.79 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.88 |
| -9999.00 |
| 0.46 |
| -9999.00 |
| -1.22 |
| -9999.00 |
| -0.05 |
| -9999.00 |
| -1.35 |
| -9999.00 |
| 0.43 |
| -9999.00 |
| -1.17 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -2.13 |
| -9999.00 |

STAR_WARN,SN_WARN
SMC6
| 3765. | +/- | 15. |
| 3866. | +/- | 94. |
| 4.42 | +/- | 0. |
| 4.82 | +/- | 0. |
| 0.75 | +/- | 0. |
| 0.75 | +/- | -NaN |
| -0.29 | +/- | 0. |
| -0.29 | +/- | 0. |
| 0.02 | +/- | 0. |
| 0.02 | +/- | -NaN |
| -0.41 | +/- | 0. |
| -0.41 | +/- | -NaN |
| -0.01 | +/- | 0. |
| -0.02 | +/- | 0. |
| 5.38 | +/- | 0. |
| 0.73 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.46 |
| -9999.00 |
| -0.42 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.30 |
| -9999.00 |
| -0.17 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| 0.15 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.30 |
| -9999.00 |
| -0.27 |
| -9999.00 |
| -0.45 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.12 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.28 |
| -9999.00 |
| -1.30 |
| -9999.00 |
STAR_WARN,SN_WARN
SMC6
| 3696. | +/- | 5. |
| 3841. | +/- | 101. |
| -0.02 | +/- | 0. |
| 0.01 | +/- | 0. |
| 2.55 | +/- | 0. |
| 2.55 | +/- | -NaN |
| -1.32 | +/- | 0. |
| -1.32 | +/- | 0. |
| -0.07 | +/- | 0. |
| -0.07 | +/- | -NaN |
| 0.26 | +/- | 0. |
| 0.26 | +/- | -NaN |
| 0.03 | +/- | 0. |
| -0.01 | +/- | 0. |
| 6.39 | +/- | 0. |
| 0.81 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.07 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -1.16 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| -1.59 |
| -9999.00 |
| 0.09 |
| -9999.00 |
| -1.15 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -1.27 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.41 |
| -9999.00 |
| -1.32 |
| -9999.00 |
| -1.64 |
| -9999.00 |
| -1.06 |
| -9999.00 |
| -1.73 |
| -9999.00 |
| -2.50 |
| -9999.00 |
| -0.89 |
| -9999.00 |
| -1.17 |
| -9999.00 |
LOW_SNR,PERSIST_JUMP_NEG
STAR_WARN,SN_WARN
SMC6
| 3986. | +/- | 6. |
| 4130. | +/- | 106. |
| 0.83 | +/- | 0. |
| 0.85 | +/- | 0. |
| 2.25 | +/- | 0. |
| 2.25 | +/- | -NaN |
| -1.30 | +/- | 0. |
| -1.30 | +/- | 0. |
| -0.56 | +/- | 0. |
| -0.56 | +/- | -NaN |
| -0.12 | +/- | 0. |
| -0.12 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.10 | +/- | 0. |
| 6.34 | +/- | 0. |
| 0.80 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.53 |
| -9999.00 |
| -1.05 |
| -9999.00 |
| -0.09 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.59 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -1.66 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| -1.51 |
| -9999.00 |
| 0.42 |
| -9999.00 |
| -1.53 |
| -9999.00 |
| -0.02 |
| -9999.00 |
| -0.26 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -1.63 |
| -9999.00 |
| -1.40 |
| -9999.00 |
| -1.56 |
| -9999.00 |
| -1.29 |
| -9999.00 |
| -1.47 |
| -9999.00 |
| -1.45 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| -0.85 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -9999.00 |
| -9999.00 |
| -2.45 |
| -9999.00 |
| -1.15 |
| -9999.00 |

SMC6
| 6347. | +/- | 17. |
| 6223. | +/- | 137. |
| 4.37 | +/- | 0. |
| 4.19 | +/- | 0. |
| 1.28 | +/- | 0. |
| 1.28 | +/- | -NaN |
| -0.05 | +/- | 0. |
| -0.04 | +/- | 0. |
| -0.15 | +/- | 0. |
| -0.15 | +/- | -NaN |
| 0.20 | +/- | 0. |
| 0.20 | +/- | -NaN |
| 0.04 | +/- | 0. |
| 0.03 | +/- | 0. |
| 7.39 | +/- | 0. |
| 0.87 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.08 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| 0.21 |
| -9999.00 |
| 0.14 |
| -9999.00 |
| -0.15 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.23 |
| -9999.00 |
| 0.13 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.11 |
| -9999.00 |
| -0.40 |
| -9999.00 |
| -0.20 |
| -9999.00 |
| -0.22 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.80 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| 0.25 |
| -9999.00 |
| -0.66 |
| -9999.00 |
| 0.24 |
| -9999.00 |
| -0.11 |
| -9999.00 |
STAR_BAD,COLORTE_BAD,SN_BAD
STAR_WARN,COLORTE_WARN,SN_WARN
SMC6
| 5434. | +/- | 327. |
| -10000. | +/- | -NaN |
| 4.20 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.98 | +/- | 1. |
| 0.98 | +/- | -NaN |
| -1.69 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| -0.03 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.09 | +/- | 1. |
| -9999.99 | +/- | -NaN |
| 0.52 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 8.90 | +/- | 0. |
| 0.95 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.04 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 1.50 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| 0.51 |
| -9999.00 |
| -1.39 |
| -9999.00 |
| 0.63 |
| -9999.00 |
| -1.70 |
| -9999.00 |
| 0.92 |
| -9999.00 |
| -0.31 |
| -9999.00 |
| -0.75 |
| -9999.00 |
| -0.65 |
| -9999.00 |
| 0.68 |
| -9999.00 |
| -0.55 |
| -9999.00 |
| -2.49 |
| -9999.00 |
| -0.99 |
| -9999.00 |
| -1.79 |
| -9999.00 |
| -1.93 |
| -9999.00 |
| -1.72 |
| -9999.00 |
| -0.29 |
| -9999.00 |
| -1.60 |
| -9999.00 |
| -0.48 |
| -9999.00 |
| -1.85 |
| -9999.00 |
| -0.01 |
| -9999.00 |
| -2.46 |
| -9999.00 |

SMC6
| 6336. | +/- | 18. |
| 6214. | +/- | 137. |
| 4.40 | +/- | 0. |
| 4.22 | +/- | 0. |
| 1.41 | +/- | 0. |
| 1.41 | +/- | -NaN |
| -0.07 | +/- | 0. |
| -0.06 | +/- | 0. |
| -0.10 | +/- | 0. |
| -0.10 | +/- | -NaN |
| 0.22 | +/- | 0. |
| 0.22 | +/- | -NaN |
| 0.03 | +/- | 0. |
| 0.02 | +/- | 0. |
| 7.09 | +/- | 0. |
| 0.85 | +/- | -NaN |
| 0.00 | +/- | 0. |
| -9999.99 | +/- | -NaN |
| 0.02 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| 0.22 |
| -9999.00 |
| 0.18 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| 0.02 |
| -9999.00 |
| 0.06 |
| -9999.00 |
| 0.01 |
| -9999.00 |
| -0.06 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| -0.21 |
| -9999.00 |
| -0.04 |
| -9999.00 |
| -0.67 |
| -9999.00 |
| -0.18 |
| -9999.00 |
| -0.00 |
| -9999.00 |
| -0.37 |
| -9999.00 |
| -0.25 |
| -9999.00 |
| -0.07 |
| -9999.00 |
| 0.04 |
| -9999.00 |
| -0.03 |
| -9999.00 |
| -0.10 |
| -9999.00 |
| -0.12 |
| -9999.00 |
| 0.20 |
| -9999.00 |
| -0.33 |
| -9999.00 |
| 0.03 |
| -9999.00 |
| 0.05 |
| -9999.00 |
